Better Epitope Discovery, Precision Immune Engineering, and Accelerated Vaccine Design Using Immunoinformatics Tools
- PMID: 32318055
- PMCID: PMC7154102
- DOI: 10.3389/fimmu.2020.00442
Better Epitope Discovery, Precision Immune Engineering, and Accelerated Vaccine Design Using Immunoinformatics Tools
Abstract
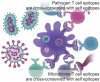
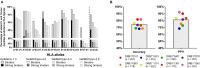
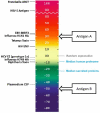
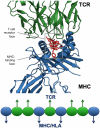
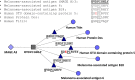
Computational vaccinology includes epitope mapping, antigen selection, and immunogen design using computational tools. Tools that facilitate the in silico prediction of immune response to biothreats, emerging infectious diseases, and cancers can accelerate the design of novel and next generation vaccines and their delivery to the clinic. Over the past 20 years, vaccinologists, bioinformatics experts, and advanced programmers based in Providence, Rhode Island, USA have advanced the development of an integrated toolkit for vaccine design called iVAX, that is secure and user-accessible by internet. This integrated set of immunoinformatic tools comprises algorithms for scoring and triaging candidate antigens, selecting immunogenic and conserved T cell epitopes, re-engineering or eliminating regulatory T cell epitopes, and re-designing antigens to induce immunogenicity and protection against disease for humans and livestock. Commercial and academic applications of iVAX have included identifying immunogenic T cell epitopes in the development of a T-cell based human multi-epitope Q fever vaccine, designing novel influenza vaccines, identifying cross-conserved T cell epitopes for a malaria vaccine, and analyzing immune responses in clinical vaccine studies. Animal vaccine applications to date have included viral infections of pigs such as swine influenza A, PCV2, and African Swine Fever. "Rapid-Fire" applications for biodefense have included a demonstration project for Lassa Fever and Q fever. As recent infectious disease outbreaks underscore the significance of vaccine-driven preparedness, the integrated set of tools available on the iVAX toolkit stand ready to help vaccine developers deliver genome-derived, epitope-driven vaccines.
Keywords: ClustiMer; EpiMatrix; JanusMatrix; T cell epitope; Treg epitope; bioinformatics; immunoinformatics; vaccines.
Copyright © 2020 De Groot, Moise, Terry, Gutierrez, Hindocha, Richard, Hoft, Ross, Noe, Takahashi, Kotraiah, Silk, Nielsen, Minassian, Ashfield, Ardito, Draper and Martin.
Figures

References
-
- DiPiazza AT, Fan S, Rattan A, DeDiego ML, Chaves F, Neumann G, et al. . A novel vaccine strategy to overcome poor immunogenicity of avian influenza vaccines through mobilization of memory CD4 T cells established by seasonal influenza. J Immunol. (2019) 203:1502–8. 10.4049/jimmunol.1900819 - DOI - PMC - PubMed
-
- Moise L, Terry F, Ardito M, Tassone R, Latimer H, Boyle C, et al. . Universal H1N1 influenza vaccine development: identification of consensus class II hemagglutinin and neuraminidase epitopes derived from strains circulating between 1980 and 2011. Hum Vaccin Immunother. (2013) 9:1598–607. 10.4161/hv.25598 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

