Divide to Conquer: Evolutionary History of Allioideae Tribes (Amaryllidaceae) Is Linked to Distinct Trends of Karyotype Evolution
- PMID: 32318079
- PMCID: PMC7155398
- DOI: 10.3389/fpls.2020.00320
Divide to Conquer: Evolutionary History of Allioideae Tribes (Amaryllidaceae) Is Linked to Distinct Trends of Karyotype Evolution
Abstract
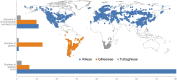
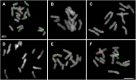
Allioideae (e.g., chives, garlics, onions) comprises three mainly temperate tribes: Allieae (800 species from the northern hemisphere), Gilliesieae (80 South American species), and Tulbaghieae (26 Southern African species). We reconstructed the phylogeny of Allioideae (190 species plus 257 species from Agapanthoideae and Amaryllidoideae) based on ITS, matK, ndhF, and rbcL to investigate its historical biogeography and karyotype evolution using newly generated cytomolecular data for Chilean Gilliesieae genera Gethyum, Miersia, Solaria, and Speea. The crown group of Allioideae diversified ∼62 Mya supporting a Gondwanic origin for the subfamily and vicariance as the cause of the intercontinental disjunction of the tribes. Our results support the hypothesis of the Indian tectonic plate carrying Allieae to northern hemisphere ('out-of-India' hypothesis). The colonization of the northern hemisphere (∼30 Mya) is correlated with a higher diversification rate in Allium associated to stable x = 8, increase of polyploidy and the geographic expansion in Europe and North America. Tulbaghieae presented x = 6, but with numerical stability (2n = 12). In contrast, the tribe Gilliesieae (x = 6) varied considerably in genome size (associated with Robertsonian translocations), rDNA sites distribution and chromosome number. Our data indicate that evolutionary history of Allioideae tribes is linked to distinct trends of karyotype evolution.
Keywords: Amaryllidaceae; BioGeoBEARS; biogeography; cytogenetics; genome size; phylogenetic comparative methods (PCMs); rDNA sites.
Copyright © 2020 Costa, Jimenez, Carvalho, Carvalho-Sobrinho, Escobar and Souza.
Figures




References
-
- Akaike H. A. (1974). New look at the statistical model identification. IEEE Trans. Automat. Contr. 19 716–723. 10.1109/TAC.1974.1100705 - DOI
-
- Angiosperm Phylogeny Group [APG] (2016). An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot. J. Linn. Soc. 181 1–20. 10.1111/boj.12385 - DOI
-
- Bartish I. V., Antonelli A., Richardson J. E., Swenson U. (2011). Vicariance or long-distance dispersal: historical biogeography of the pantropical subfamily chrysophylloideae (Sapotaceae). J. Biogeogr. 38 177–190. 10.1111/j.1365-2699.2010.02389.x - DOI
LinkOut - more resources
Full Text Sources
Miscellaneous

