Integrated analysis of transcriptomic and metabolomic profiling reveal the p53 associated pathways underlying the response to ionizing radiation in HBE cells
- PMID: 32318262
- PMCID: PMC7160934
- DOI: 10.1186/s13578-020-00417-z
Integrated analysis of transcriptomic and metabolomic profiling reveal the p53 associated pathways underlying the response to ionizing radiation in HBE cells
Abstract
Background: Radiation damage to normal tissues is a serious concern. P53 is a well-known transcription factor which is closely associated with radiation-induced cell damage. Increasing evidence has indicated that regulation of metabolism by p53 represents a reviving mechanism vital to protect cell survival. We aimed to explore the interactions of radiation-induced transcripts with the cellular metabolism regulated by p53.
Methods: Human bronchial epithelial (HBE) cell line was used to knockout p53 using CRISPR/cas9. Transcriptomic analysis was conducted by microarray and metabolomic analysis was conducted by GC-MS. Integrative omics was performed using MetaboAnalyst.
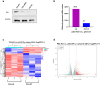
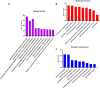
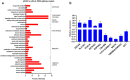
Results: 326 mRNAs showed significantly altered expression in HBE p53-/- cells post-radiation, of which 269 were upregulated and 57 were downregulated. A total of 147 metabolites were altered, including 45 that increased and 102 that decreased. By integrated analysis of both omic data, we found that in response to radiation insult, nitrogen metabolism, glutathione metabolism, arachidonic acid metabolism, and glycolysis or gluconeogenesis may be dysregulated due to p53.
Conclusions: Our study provided a pilot comprehensive view of the metabolism regulated by p53 in response to radiation exposure. Detailed evaluation of these important p53-regulated metabolic pathways, including their roles in the response to radiation of cells, is essential to elucidate the molecular mechanisms of radiation-induced damage.
© The Author(s) 2020.
Conflict of interest statement
Competing interestsThe authors declare no competing financial interests.
Figures



References
-
- MacVittie TJ, Bennett AW, Farese AM, Taylor-Howell C, Smith CP, Gibbs AM, et al. The effect of radiation dose and variation in neupogen (R) initiation schedule on the mitigation of myelosuppression during the concomitant GI-ARS and H-ARS in a nonhuman primate model of high-dose exposure with marrow sparing. Health Phys. 2015;109:427–439. doi: 10.1097/HP.0000000000000350. - DOI - PMC - PubMed
-
- MacVittie TJ, Farese AM, Kane MA. ARS, DEARE, and multiple-organ injury: a strategic and tactical approach to link radiation effects, animal models, medical countermeasures, and biomarker development to predict clinical outcome. Health Phys. 2019;116:453. doi: 10.1097/HP.0000000000001050. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

