Computational Oncology in the Multi-Omics Era: State of the Art
- PMID: 32318338
- PMCID: PMC7154096
- DOI: 10.3389/fonc.2020.00423
Computational Oncology in the Multi-Omics Era: State of the Art
Abstract
Cancer is the quintessential complex disease. As technologies evolve faster each day, we are able to quantify the different layers of biological elements that contribute to the emergence and development of malignancies. In this multi-omics context, the use of integrative approaches is mandatory in order to gain further insights on oncological phenomena, and to move forward toward the precision medicine paradigm. In this review, we will focus on computational oncology as an integrative discipline that incorporates knowledge from the mathematical, physical, and computational fields to further the biomedical understanding of cancer. We will discuss the current roles of computation in oncology in the context of multi-omic technologies, which include: data acquisition and processing; data management in the clinical and research settings; classification, diagnosis, and prognosis; and the development of models in the research setting, including their use for therapeutic target identification. We will discuss the machine learning and network approaches as two of the most promising emerging paradigms, in computational oncology. These approaches provide a foundation on how to integrate different layers of biological description into coherent frameworks that allow advances both in the basic and clinical settings.
Keywords: cancer complexity; computational oncology; data integration; machine learning; multi-omics analysis; network science.
Copyright © 2020 de Anda-Jáuregui and Hernández-Lemus.
Figures
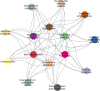


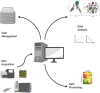

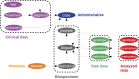
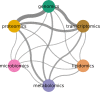

References
-
- Sayama H. Introduction to the Modeling and Analysis of Complex Systems. Geneseo, NY: Open SUNY Textbooks (2015). Available online at: http://textbooks.opensuny.org/introduction-to-the-modeling-and-analysis-...
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

