CytoMAP: A Spatial Analysis Toolbox Reveals Features of Myeloid Cell Organization in Lymphoid Tissues
- PMID: 32320656
- PMCID: PMC7233132
- DOI: 10.1016/j.celrep.2020.107523
CytoMAP: A Spatial Analysis Toolbox Reveals Features of Myeloid Cell Organization in Lymphoid Tissues
Abstract
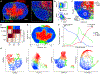
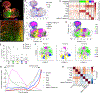
Recently developed approaches for highly multiplexed imaging have revealed complex patterns of cellular positioning and cell-cell interactions with important roles in both cellular- and tissue-level physiology. However, tools to quantitatively study cellular patterning and tissue architecture are currently lacking. Here, we develop a spatial analysis toolbox, the histo-cytometric multidimensional analysis pipeline (CytoMAP), which incorporates data clustering, positional correlation, dimensionality reduction, and 2D/3D region reconstruction to identify localized cellular networks and reveal features of tissue organization. We apply CytoMAP to study the microanatomy of innate immune subsets in murine lymph nodes (LNs) and reveal mutually exclusive segregation of migratory dendritic cells (DCs), regionalized compartmentalization of SIRPα- dermal DCs, and preferential association of resident DCs with select LN vasculature. The findings provide insights into the organization of myeloid cells in LNs and demonstrate that CytoMAP is a comprehensive analytics toolbox for revealing features of tissue organization in imaging datasets.
Keywords: cellular organization; dendritic cell positioning; lymphoid tissue anatomy; machine learning; quantitative image analysis; quantitative microscopy; spatial analysis; tissue microanatomy; tissue organization; tumor microenvironments.
Copyright © 2020 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests M.P. and T.P. are employees of Roche. All other authors declare no competing interests.
Figures





References
-
- Baptista AP, Gola A, Huang Y, Milanez-Almeida P, Torabi-Parizi P, Urban JF Jr., Shapiro VS, Gerner MY, and Germain RN (2019). The chemoattractant receptor Ebi2 drives intranodal naive CD4+ T cell peripheralization to promote effective adaptive immunity. Immunity 50, 1188–1201.e6. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

