Emerging SARS-CoV-2 mutation hot spots include a novel RNA-dependent-RNA polymerase variant
- PMID: 32321524
- PMCID: PMC7174922
- DOI: 10.1186/s12967-020-02344-6
Emerging SARS-CoV-2 mutation hot spots include a novel RNA-dependent-RNA polymerase variant
Abstract
Background: SARS-CoV-2 is a RNA coronavirus responsible for the pandemic of the Severe Acute Respiratory Syndrome (COVID-19). RNA viruses are characterized by a high mutation rate, up to a million times higher than that of their hosts. Virus mutagenic capability depends upon several factors, including the fidelity of viral enzymes that replicate nucleic acids, as SARS-CoV-2 RNA dependent RNA polymerase (RdRp). Mutation rate drives viral evolution and genome variability, thereby enabling viruses to escape host immunity and to develop drug resistance.
Methods: We analyzed 220 genomic sequences from the GISAID database derived from patients infected by SARS-CoV-2 worldwide from December 2019 to mid-March 2020. SARS-CoV-2 reference genome was obtained from the GenBank database. Genomes alignment was performed using Clustal Omega. Mann-Whitney and Fisher-Exact tests were used to assess statistical significance.
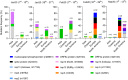
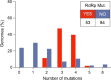
Results: We characterized 8 novel recurrent mutations of SARS-CoV-2, located at positions 1397, 2891, 14408, 17746, 17857, 18060, 23403 and 28881. Mutations in 2891, 3036, 14408, 23403 and 28881 positions are predominantly observed in Europe, whereas those located at positions 17746, 17857 and 18060 are exclusively present in North America. We noticed for the first time a silent mutation in RdRp gene in England (UK) on February 9th, 2020 while a different mutation in RdRp changing its amino acid composition emerged on February 20th, 2020 in Italy (Lombardy). Viruses with RdRp mutation have a median of 3 point mutations [range: 2-5], otherwise they have a median of 1 mutation [range: 0-3] (p value < 0.001).
Conclusions: These findings suggest that the virus is evolving and European, North American and Asian strains might coexist, each of them characterized by a different mutation pattern. The contribution of the mutated RdRp to this phenomenon needs to be investigated. To date, several drugs targeting RdRp enzymes are being employed for SARS-CoV-2 infection treatment. Some of them have a predicted binding moiety in a SARS-CoV-2 RdRp hydrophobic cleft, which is adjacent to the 14408 mutation we identified. Consequently, it is important to study and characterize SARS-CoV-2 RdRp mutation in order to assess possible drug-resistance viral phenotypes. It is also important to recognize whether the presence of some mutations might correlate with different SARS-CoV-2 mortality rates.
Keywords: COVID-19; Drug resistance; Europe; Mutation; Pneumonia; RNA-dependent-RNA-polymerase; RdRp; SARS-CoV-2; Viral mutagenesis.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

