Isolation and characterization of diverse microbial representatives from the human skin microbiome
- PMID: 32321582
- PMCID: PMC7178971
- DOI: 10.1186/s40168-020-00831-y
Isolation and characterization of diverse microbial representatives from the human skin microbiome
Abstract
Background: The skin micro-environment varies across the body, but all sites are host to microorganisms that can impact skin health. Some of these organisms are true commensals which colonize a unique niche on the skin, while open exposure of the skin to the environment also results in the transient presence of diverse microbes with unknown influences on skin health. Culture-based studies of skin microbiota suggest that skin microbes can affect skin properties, immune responses, pathogen growth, and wound healing.
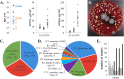
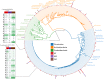
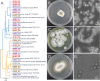
Results: In this work, we greatly expanded the diversity of available commensal organisms by collecting > 800 organisms from 3 body sites of 17 individuals. Our collection includes > 30 bacterial genera and 14 fungal genera, with Staphylococcus and Micrococcus as the most prevalent isolates. We characterized a subset of skin isolates for the utilization of carbon compounds found on the skin surface. We observed that members of the skin microbiota have the capacity to metabolize amino acids, steroids, lipids, and sugars, as well as compounds originating from personal care products.
Conclusions: This collection is a resource that will support skin microbiome research with the potential for discovery of novel small molecules, development of novel therapeutics, and insight into the metabolic activities of the skin microbiota. We believe this unique resource will inform skin microbiome management to benefit skin health. Video abstract.
Keywords: Carbon source utilization; Isolate collection; Skin microbiome.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

