Robust Summarization and Inference in Proteome-wide Label-free Quantification
- PMID: 32321741
- PMCID: PMC7338080
- DOI: 10.1074/mcp.RA119.001624
Robust Summarization and Inference in Proteome-wide Label-free Quantification
Abstract
Label-Free Quantitative mass spectrometry based workflows for differential expression (DE) analysis of proteins impose important challenges on the data analysis because of peptide-specific effects and context dependent missingness of peptide intensities. Peptide-based workflows, like MSqRob, test for DE directly from peptide intensities and outperform summarization methods which first aggregate MS1 peptide intensities to protein intensities before DE analysis. However, these methods are computationally expensive, often hard to understand for the non-specialized end-user, and do not provide protein summaries, which are important for visualization or downstream processing. In this work, we therefore evaluate state-of-the-art summarization strategies using a benchmark spike-in dataset and discuss why and when these fail compared with the state-of-the-art peptide based model, MSqRob. Based on this evaluation, we propose a novel summarization strategy, MSqRobSum, which estimates MSqRob's model parameters in a two-stage procedure circumventing the drawbacks of peptide-based workflows. MSqRobSum maintains MSqRob's superior performance, while providing useful protein expression summaries for plotting and downstream analysis. Summarizing peptide to protein intensities considerably reduces the computational complexity, the memory footprint and the model complexity, and makes it easier to disseminate DE inferred on protein summaries. Moreover, MSqRobSum provides a highly modular analysis framework, which provides researchers with full flexibility to develop data analysis workflows tailored toward their specific applications.
Keywords: Biostatistics; bioinformatics; bioinformatics software; differential expression; label-free quantification; mass spectrometry; ridge regression; shotgun proteomics; summarization.
© 2020 Sticker et al.
Conflict of interest statement
Conflict of interest—Authors declare no competing interests.
Figures

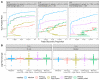


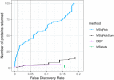
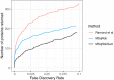
References
-
- Goeminne L. J. E., Gevaert K., and Clement L. (2018) Experimental design and data-analysis in label-free quantitative LC/MS proteomics: A tutorial with MSqRob. J. Proteomics 171, 23–36 - PubMed
-
- Tebbe A., Klammer M., Sighart S., Schaab C., and Daub H. (2015) Systematic evaluation of label-free and super-SILAC quantification for proteome expression analysis. Rapid Commun. Mass Spectrom. 29, 795–801 - PubMed
-
- Lazar C., Gatto L., Ferro M., Bruley C., and Burger T. (2016) Accounting for the multiple natures of missing values in label-free quantitative proteomics data sets to compare imputation strategies. J. Proteome Res. 15, 1116–1125 - PubMed
-
- Goeminne L.J.E., Argentini A., Martens L., and Clement L. (2015) Summarization vs peptide-based models in label-free quantitative proteomics: performance, pitfalls, and data analysis guidelines. J. Proteome Res. 14, 2457–2465 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

