Cyclophilin A Inhibitor Debio-025 Targets Crk, Reduces Metastasis, and Induces Tumor Immunogenicity in Breast Cancer
- PMID: 32321766
- PMCID: PMC8045419
- DOI: 10.1158/1541-7786.MCR-19-1144
Cyclophilin A Inhibitor Debio-025 Targets Crk, Reduces Metastasis, and Induces Tumor Immunogenicity in Breast Cancer
Abstract
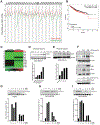
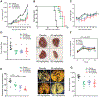
The Crk adaptor protein, a critical modifier of multiple signaling pathways, is overexpressed in many cancers where it contributes to tumor progression and metastasis. Recently, we have shown that Crk interacts with the peptidyl prolyl cis-trans isomerase, Cyclophilin A (CypA; PP1A) via a G219P220Y221 (GPY) motif in the carboxyl-terminal linker region of Crk, thereby delaying pY221 phosphorylation and preventing downregulation of Crk signaling. Here, we investigate the physiologic significance of the CypA/Crk interaction and query whether CypA inhibition affects Crk signaling in vitro and in vivo. We show that CypA, when induced under conditions of hypoxia, regulates Crk pY221 phosphorylation and signaling in cancer cell lines. Using nuclear magnetic resonance spectroscopy, we show that CypA binds to the Crk GPY motif via the catalytic PPII domain of CypA, and small-molecule nonimmunosuppressive inhibitors of CypA (Debio-025) disrupt the CypA-CrkII interaction and restores phosphorylation of Crk Y221. In cultured cell lines, Debio-025 suppresses cell migration, and when administered in vivo in an orthotopic model of triple-negative breast cancer, Debio-025 showed antitumor efficacy either alone or in combination with anti-PD-1 mAb, reducing both tumor volume and metastatic lung dispersion. Furthermore, when analyzed by NanoString immune profiling, treatment of Debio-025 with anti-PD-1 mAb increased both T-cell signaling and innate immune signaling in tumor microenvironment. IMPLICATIONS: These data suggest that pharmacologic inhibition of CypA may provide a promising and unanticipated consequence in cancer biology, in part by targeting the CypA/CrkII axis that regulates cell migration, tumor metastasis, and host antitumor immune evasion.
©2020 American Association for Cancer Research.
Conflict of interest statement
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Figures




References
-
- Matsuda M, Mayer BJ, Fukui Y, Hanafusa H. Binding of transforming protein, P47gag-crk, to a broad range of phosphotyrosine-containing proteins. Science 1990;248:1537–9. - PubMed
-
- Mayer BJ, Hamaguchi M, Hanafusa H. A novel viral oncogene with structural similarity to phospholipase C. Nature 1988;332:272–5. - PubMed
-
- Hanafusa H Protein phosphorylation and cell transformation. Zhonghua Min Guo Wei Sheng Wu Ji Mian Yi Xue Za Zhi 1991;24:1–9. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

