Impact of HLA-B*52:01-Driven Escape Mutations on Viral Replicative Capacity
- PMID: 32321820
- PMCID: PMC7307159
- DOI: 10.1128/JVI.02025-19
Impact of HLA-B*52:01-Driven Escape Mutations on Viral Replicative Capacity
Abstract
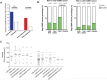
HLA-B*52:01 is strongly associated with protection against HIV disease progression. However, the mechanisms of HLA-B*52:01-mediated immune control have not been well studied. We here describe a cohort with a majority of HIV C-clade-infected individuals from Delhi, India, where HLA-B*52:01 is highly prevalent (phenotypic frequency, 22.5%). Consistent with studies of other cohorts, expression of HLA-B*52:01 was associated with high absolute CD4 counts and therefore a lack of HIV disease progression. We here examined the impact of HLA-B*52:01-associated viral polymorphisms within the immunodominant C clade Gag epitope RMTSPVSI (here, RI8; Gag residues 275 to 282) on viral replicative capacity (VRC) since HLA-mediated reduction in VRC is a central mechanism implicated in HLA-associated control of HIV. We observed in HLA-B*52:01-positive individuals a higher frequency of V280T, V280S, and V280A variants within RI8 (P = 0.0001). Each of these variants reduced viral replicative capacity in C clade viruses, particularly the V280A variant (P < 0.0001 in both the C clade consensus and in the Indian study cohort consensus p24 Gag backbone), which was also associated with significantly higher absolute CD4 counts in the donors (median, 941.5 cells/mm3; P = 0.004). A second HLA-B*52:01-associated mutation, K286R, flanking HLA-B*52:01-RI8, was also analyzed. Although selected in HLA-B*52:01-positive subjects often in combination with the V280X variants, this mutation did not act as a compensatory mutant but, indeed, further reduced VRC. These data are therefore consistent with previous work showing that HLA-B molecules that are associated with immune control of HIV principally target conserved epitopes within the capsid protein, escape from which results in a significant reduction in VRC.IMPORTANCE Few studies have addressed the mechanisms of immune control in HIV-infected subjects in India, where an estimated 2.7 million people are living with HIV. We focus here on a study cohort in Delhi on one of the most prevalent HLA-B alleles, HLA-B*52:01, present in 22.5% of infected individuals. HLA-B*52:01 has consistently been shown in other cohorts to be associated with protection against HIV disease progression, but studies have been limited by the low prevalence of this allele in North America and Europe. Among the C-clade-infected individuals, we show that HLA-B*52:01 is the most protective of all the HLA-B alleles expressed in the Indian cohort and is associated with the highest absolute CD4 counts. Further, we show that the mechanism by which HLA-B*52:01 mediates immune protection is, at least in part, related to the inability of HIV to evade the HLA-B*52:01-restricted p24 Gag-specific CD8+ T-cell response without incurring a significant loss to viral replicative capacity.
Keywords: C clade; CTL; HLA; HLA-B*52:01; Indian; compensatory mutation; escape mutation; human immunodeficiency virus; p24 Gag; viral replicative capacity.
Copyright © 2020 American Society for Microbiology.
Figures



Similar articles
-
Consequences of HLA-B*13-Associated Escape Mutations on HIV-1 Replication and Nef Function.J Virol. 2015 Nov;89(22):11557-71. doi: 10.1128/JVI.01955-15. Epub 2015 Sep 9. J Virol. 2015. PMID: 26355081 Free PMC article.
-
Differential escape patterns within the dominant HLA-B*57:03-restricted HIV Gag epitope reflect distinct clade-specific functional constraints.J Virol. 2014 May;88(9):4668-78. doi: 10.1128/JVI.03303-13. Epub 2014 Feb 5. J Virol. 2014. PMID: 24501417 Free PMC article.
-
Pol-Driven Replicative Capacity Impacts Disease Progression in HIV-1 Subtype C Infection.J Virol. 2018 Sep 12;92(19):e00811-18. doi: 10.1128/JVI.00811-18. Print 2018 Oct 1. J Virol. 2018. PMID: 29997209 Free PMC article.
-
The influence of HLA/HIV genetics on the occurrence of elite controllers and a need for therapeutics geotargeting view.Braz J Infect Dis. 2021 Sep-Oct;25(5):101619. doi: 10.1016/j.bjid.2021.101619. Epub 2021 Sep 22. Braz J Infect Dis. 2021. PMID: 34562387 Free PMC article. Review.
-
Characterization of the Protective HIV-1 CTL Epitopes and the Corresponding HLA Class I Alleles: A Step towards Designing CTL Based HIV-1 Vaccine.Adv Virol. 2014;2014:321974. doi: 10.1155/2014/321974. Epub 2014 Mar 18. Adv Virol. 2014. PMID: 24744786 Free PMC article. Review.
Cited by
-
Effect of Difference in Consensus Sequence between HIV-1 Subtype A/E and Subtype B Viruses on Elicitation of Gag-Specific CD8+ T Cells and Accumulation of HLA-Associated Escape Mutations.J Virol. 2021 Feb 24;95(6):e02061-20. doi: 10.1128/JVI.02061-20. Print 2021 Feb 24. J Virol. 2021. PMID: 33361435 Free PMC article.
-
Computational and Population-Based HLA Permissiveness to HIV Drug Resistance-Associated Mutations.Pathogens. 2025 Feb 20;14(3):207. doi: 10.3390/pathogens14030207. Pathogens. 2025. PMID: 40137693 Free PMC article.
-
Definition of a New HLA B*52-Restricted Rev CTL Epitope Targeted by an HIV-1-Infected Controller.Viruses. 2023 Feb 18;15(2):567. doi: 10.3390/v15020567. Viruses. 2023. PMID: 36851781 Free PMC article.
References
-
- Kiepiela P, Leslie A, Honeyborne I, Ramduth D, Thobakgale C, Chetty S, Rathnavalu P, Moore C, Pfafferott K, Hilton L, Zimbwa P, Moore S, Allen T, Brander C, Addo M, Altfeld M, James I, Mallal S, Bunce M, Barber L, Szinger J, Day C, Klenerman P, Mullins J, Korber B, Coovadia H, Walker B, Goulder P. 2004. Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA. Nature 432:769–775. doi:10.1038/nature03113. - DOI - PubMed
-
- International HIV Controllers Study, Pereyra F, Jia X, McLaren PJ, Telenti A, deBakker PI, Walker BD, Ripke S, Brumme CJ, Pulit SL, Carrington M, Kadie CM, Carlson JM, Heckerman D, Graham RR, Plenge RM, Deeks SG, Gianniny L, Crawford G, Sullivan J, Gonzalez E, Davies L, Camargo A, Moore JM, Beattie N, Gupta S, Crenshaw A, Burtt NP, Guiducci C, Gupta N, Gao X, Qi Y, Yuki Y, Piechocka-Trocha A, Cutrell E, Rosenberg R, Moss KL, Lemay P, O’Leary J, Schaefer T, Verma P, Toth I, Block B, Baker B, Rothchild A, Lian J, Proudfoot J, Alvino DM, Vine S, Addo MM, Allen TM, Altfeld M, et al. . 2010. The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science 330:1551–1557. doi:10.1126/science.1195271. - DOI - PMC - PubMed
-
- Antoni G, Guergnon J, Meaudre C, Samri A, Boufassa F, Goujard C, Lambotte O, Autran B, Rouzioux C, Costagliola D, Meyer L, Theodorou I. 2013. MHC-driven HIV-1 control on the long run is not systematically determined at early times post-HIV-1 infection. AIDS 27:1707–1716. doi:10.1097/QAD.0b013e328360a4bd. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

