A high definition picture of somatic mutations in chronic lymphoproliferative disorder of natural killer cells
- PMID: 32321919
- PMCID: PMC7176632
- DOI: 10.1038/s41408-020-0309-2
A high definition picture of somatic mutations in chronic lymphoproliferative disorder of natural killer cells
Abstract
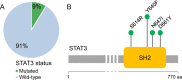
The molecular pathogenesis of chronic lymphoproliferative disorder of natural killer (NK) cells (CLPD-NK) is poorly understood. Following the screening of 57 CLPD-NK patients, only five presented STAT3 mutations. WES profiling of 13 cases negative for STAT3/STAT5B mutations uncovered an average of 18 clonal, population rare and deleterious somatic variants per patient. The mutational landscape of CLPD-NK showed that most patients carry a heavy mutational burden, with major and subclonal deleterious mutations co-existing in the leukemic clone. Somatic mutations hit genes wired to cancer proliferation, survival, and migration pathways, in the first place Ras/MAPK, PI3K-AKT, in addition to JAK/STAT (PIK3R1 and PTK2). We confirmed variants with putative driver role of MAP10, MPZL1, RPS6KA1, SETD1B, TAOK2, TMEM127, and TNFRSF1A genes, and of genes linked to viral infections (DDX3X and RSF1) and DNA repair (PAXIP1). A truncating mutation of the epigenetic regulator TET2 and a variant likely abrogating PIK3R1-negative regulatory activity were validated. This study significantly furthered the view of the genes and pathways involved in CLPD-NK, indicated similarities with aggressive diseases of NK cells and detected mutated genes targetable by approved drugs, being a step forward to personalized precision medicine for CLPD-NK patients.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
- M-IMM/Sigrid Juséliuksen Säätiö (Sigrid Jusélius Foundation)/International
- IG 20216/Associazione Italiana per la Ricerca sul Cancro (Italian Association for Cancer Research)/International
- IG 20052/Associazione Italiana per la Ricerca sul Cancro (Italian Association for Cancer Research)/International
- PREMED-AL and CELL/Fondazione Cassa di Risparmio di Padova e Rovigo (Foundation Cariparo)/International
- 2017PPS2X4_003/Ministero dell'Istruzione, dell'Università e della Ricerca (Ministry of Education, University and Research)/International
LinkOut - more resources
Full Text Sources
Miscellaneous

