Establishment of macaque trophoblast stem cell lines derived from cynomolgus monkey blastocysts
- PMID: 32321940
- PMCID: PMC7176671
- DOI: 10.1038/s41598-020-63602-7
Establishment of macaque trophoblast stem cell lines derived from cynomolgus monkey blastocysts
Abstract
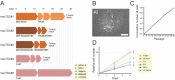
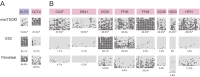
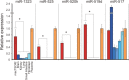
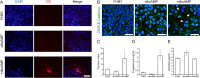
The placenta forms a maternal-fetal junction that supports many physiological functions such as the supply of nutrition and exchange of gases and wastes. Establishing an in vitro culture model of human and non-human primate trophoblast stem/progenitor cells is important for investigating the process of early placental development and trophoblast differentiation. In this study, we have established five trophoblast stem cell (TSC) lines from cynomolgus monkey blastocysts, named macTSC #1-5. Fibroblast growth factor 4 (FGF4) enhanced proliferation of macTSCs, while other exogenous factors were not required to maintain their undifferentiated state. macTSCs showed a trophoblastic gene expression profile and trophoblast-like DNA methylation status and also exhibited differentiation capacity towards invasive trophoblast cells and multinucleated syncytia. In a xenogeneic chimera assay, these stem cells contributed to trophectoderm (TE) development in the chimeric blastocysts. macTSC are the first primate trophoblast cell lines whose proliferation is promoted by FGF4. These cell lines provide a valuable in vitro culture model to analyze the similarities and differences in placental development between human and non-human primates.
Conflict of interest statement
The authors declare no competing interests.
Figures

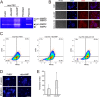

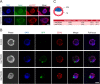
References
-
- Weber M, Knoefler I, Schleussner E, Markert UR, Fitzgerald JS. HTR8/SVneo cells display trophoblast progenitor cell-like characteristics indicative of self-renewal, repopulation activity, and expression of “stemness-” associated transcription factors. Biomed Res. Int. 2013;2013:1–10. doi: 10.1155/2013/243649. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

