Modelling the COVID-19 epidemic and implementation of population-wide interventions in Italy
- PMID: 32322102
- PMCID: PMC7175834
- DOI: 10.1038/s41591-020-0883-7
Modelling the COVID-19 epidemic and implementation of population-wide interventions in Italy
Abstract
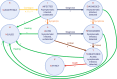
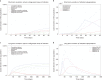
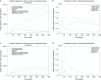
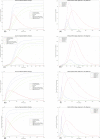
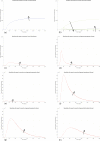
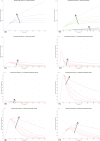
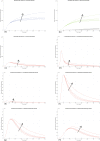
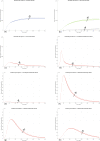
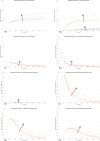
In Italy, 128,948 confirmed cases and 15,887 deaths of people who tested positive for SARS-CoV-2 were registered as of 5 April 2020. Ending the global SARS-CoV-2 pandemic requires implementation of multiple population-wide strategies, including social distancing, testing and contact tracing. We propose a new model that predicts the course of the epidemic to help plan an effective control strategy. The model considers eight stages of infection: susceptible (S), infected (I), diagnosed (D), ailing (A), recognized (R), threatened (T), healed (H) and extinct (E), collectively termed SIDARTHE. Our SIDARTHE model discriminates between infected individuals depending on whether they have been diagnosed and on the severity of their symptoms. The distinction between diagnosed and non-diagnosed individuals is important because the former are typically isolated and hence less likely to spread the infection. This delineation also helps to explain misperceptions of the case fatality rate and of the epidemic spread. We compare simulation results with real data on the COVID-19 epidemic in Italy, and we model possible scenarios of implementation of countermeasures. Our results demonstrate that restrictive social-distancing measures will need to be combined with widespread testing and contact tracing to end the ongoing COVID-19 pandemic.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- WHO. Coronavirus Disease 2019 (COVID-19): Situation Report 76 (WHO, 2020).
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

