Microbiotyping the Sinonasal Microbiome
- PMID: 32322561
- PMCID: PMC7156599
- DOI: 10.3389/fcimb.2020.00137
Microbiotyping the Sinonasal Microbiome
Abstract
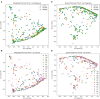
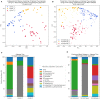
This study offers a novel description of the sinonasal microbiome, through an unsupervised machine learning approach combining dimensionality reduction and clustering. We apply our method to the International Sinonasal Microbiome Study (ISMS) dataset of 410 sinus swab samples. We propose three main sinonasal "microbiotypes" or "states": the first is Corynebacterium-dominated, the second is Staphylococcus-dominated, and the third dominated by the other core genera of the sinonasal microbiome (Streptococcus, Haemophilus, Moraxella, and Pseudomonas). The prevalence of the three microbiotypes studied did not differ between healthy and diseased sinuses, but differences in their distribution were evident based on geography. We also describe a potential reciprocal relationship between Corynebacterium species and Staphylococcus aureus, suggesting that a certain microbial equilibrium between various players is reached in the sinuses. We validate our approach by applying it to a separate 16S rRNA gene sequence dataset of 97 sinus swabs from a different patient cohort. Sinonasal microbiotyping may prove useful in reducing the complexity of describing sinonasal microbiota. It may drive future studies aimed at modeling microbial interactions in the sinuses and in doing so may facilitate the development of a tailored patient-specific approach to the treatment of sinus disease in the future.
Keywords: 16S rRNA gene; chronic rhinosinusitis; microbiome; microbiotype; next-generation sequencing; paranasal sinuses; sinus.
Copyright © 2020 Bassiouni, Paramasivan, Shiffer, Dillon, Cope, Cooksley, Ramezanpour, Moraitis, Ali, Bleier, Callejas, Cornet, Douglas, Dutra, Georgalas, Harvey, Hwang, Luong, Schlosser, Tantilipikorn, Tewfik, Vreugde, Wormald, Caporaso and Psaltis.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

