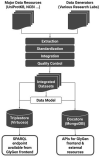
GlyGen data model and processing workflow
- PMID: 32324859
- PMCID: PMC7320628
- DOI: 10.1093/bioinformatics/btaa238
GlyGen data model and processing workflow
Abstract
Summary: Glycoinformatics plays a major role in glycobiology research, and the development of a comprehensive glycoinformatics knowledgebase is critical. This application note describes the GlyGen data model, processing workflow and the data access interfaces featuring programmatic use case example queries based on specific biological questions. The GlyGen project is a data integration, harmonization and dissemination project for carbohydrate and glycoconjugate-related data retrieved from multiple international data sources including UniProtKB, GlyTouCan, UniCarbKB and other key resources.
Availability and implementation: GlyGen web portal is freely available to access at https://glygen.org. The data portal, web services, SPARQL endpoint and GitHub repository are also freely available at https://data.glygen.org, https://api.glygen.org, https://sparql.glygen.org and https://github.com/glygener, respectively. All code is released under license GNU General Public License version 3 (GNU GPLv3) and is available on GitHub https://github.com/glygener. The datasets are made available under Creative Commons Attribution 4.0 International (CC BY 4.0) license.
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author(s) 2020. Published by Oxford University Press.
Figures