Genomic diversity of the human pathogen Paracoccidioides across the South American continent
- PMID: 32325168
- PMCID: PMC7385733
- DOI: 10.1016/j.fgb.2020.103395
Genomic diversity of the human pathogen Paracoccidioides across the South American continent
Abstract
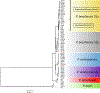
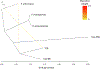
Paracoccidioidomycosis (PCM) is a life-threatening systemic mycosis widely reported in the Gran Chaco ecosystem. The disease is caused by different species from the genus Paracoccidioides, which are all endemic to South and Central America. Here, we sequenced and analyzed 31 isolates of Paracoccidioides across South America, with particular focus on isolates from Argentina and Paraguay. The de novo sequenced isolates were compared with publicly available genomes. Phylogenetics and population genomics revealed that PCM in Argentina and Paraguay is caused by three distinct Paracoccidioides genotypes, P. brasiliensis (S1a and S1b) and P. restrepiensis (PS3). P. brasiliensis S1a isolates from Argentina are frequently associated with chronic forms of the disease. Our results suggest the existence of extensive molecular polymorphism among Paracoccidioides species, and provide a framework to begin to dissect the connection between genotypic differences in the pathogen and the clinical outcomes of the disease.
Keywords: Gran Chaco; Paracoccidioides; Phylogenomics; Population genetics.
Copyright © 2020. Published by Elsevier Inc.
Conflict of interest statement
Declaration of Competing Interest The authors declare no conflicts of interest.
Figures


References
-
- Araujo P, et al., 2016. Paracoccidioidomicosis detectados en el período 2004–2013 en el Laboratorio Central de Salud Pública de Asunción - Paraguay. Revista del Nacional 8, 62–71.
-
- Bicalho RN, et al., 2001. Oral paracoccidioidomycosis: a retrospective study of 62 Brazilian patients. Oral Dis. 7, 56–60. - PubMed
-
- Brasier C, 2000. The rise of the hybrid fungi. Nature 405, 134–135. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

