Key Vitamin D Target Genes with Functions in the Immune System
- PMID: 32325790
- PMCID: PMC7230898
- DOI: 10.3390/nu12041140
Key Vitamin D Target Genes with Functions in the Immune System
Abstract
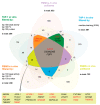
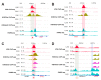
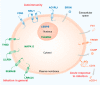
The biologically active form of vitamin D3, 1α,25-dihydroxyvitamin D3 (1,25(OH)2D3), modulates innate and adaptive immunity via genes regulated by the transcription factor vitamin D receptor (VDR). In order to identify the key vitamin D target genes involved in these processes, transcriptome-wide datasets were compared, which were obtained from a human monocytic cell line (THP-1) and peripheral blood mononuclear cells (PBMCs) treated in vitro by 1,25(OH)2D3, filtered using different approaches, as well as from PBMCs of individuals supplemented with a vitamin D3 bolus. The led to the genes ACVRL1, CAMP, CD14, CD93, CEBPB, FN1, MAPK13, NINJ1, LILRB4, LRRC25, SEMA6B, SRGN, THBD, THEMIS2 and TREM1. Public epigenome- and transcriptome-wide data from THP-1 cells were used to characterize these genes based on the level of their VDR-driven enhancers as well as the level of the dynamics of their mRNA production. Both types of datasets allowed the categorization of the vitamin D target genes into three groups according to their role in (i) acute response to infection, (ii) infection in general and (iii) autoimmunity. In conclusion, 15 genes were identified as major mediators of the action of vitamin D in innate and adaptive immunity and their individual functions are explained based on different gene regulatory scenarios.
Keywords: PBMCs; VDR; Vitamin D; epigenome; gene regulation; immune system; monocytes; transcriptome; vitamin D target genes.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Holick M.F. Photobiology of Vitamin D. Vitamin D. 2018:45–55. doi: 10.1016/B978-0-12-809965-0.00004-5. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

