Asymmetric assembly of centromeres epigenetically regulates stem cell fate
- PMID: 32328637
- PMCID: PMC7147107
- DOI: 10.1083/jcb.201910084
Asymmetric assembly of centromeres epigenetically regulates stem cell fate
Abstract
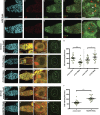
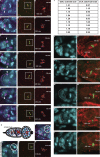
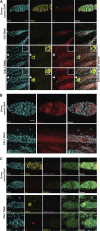
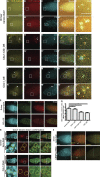
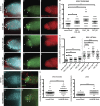
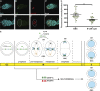
Centromeres are epigenetically defined by CENP-A-containing chromatin and are essential for cell division. Previous studies suggest asymmetric inheritance of centromeric proteins upon stem cell division; however, the mechanism and implications of selective chromosome segregation remain unexplored. We show that Drosophila female germline stem cells (GSCs) and neuroblasts assemble centromeres after replication and before segregation. Specifically, CENP-A deposition is promoted by CYCLIN A, while excessive CENP-A deposition is prevented by CYCLIN B, through the HASPIN kinase. Furthermore, chromosomes inherited by GSCs incorporate more CENP-A, making stronger kinetochores that capture more spindle microtubules and bias segregation. Importantly, symmetric incorporation of CENP-A on sister chromatids via HASPIN knockdown or overexpression of CENP-A, either alone or together with its assembly factor CAL1, drives stem cell self-renewal. Finally, continued CENP-A assembly in differentiated cells is nonessential for egg development. Our work shows that centromere assembly epigenetically drives GSC maintenance and occurs before oocyte meiosis.
© 2020 Dattoli et al.
Figures






References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

