RNA demethylase ALKBH5 promotes ovarian carcinogenesis in a simulated tumour microenvironment through stimulating NF-κB pathway
- PMID: 32329191
- PMCID: PMC7294121
- DOI: 10.1111/jcmm.15228
RNA demethylase ALKBH5 promotes ovarian carcinogenesis in a simulated tumour microenvironment through stimulating NF-κB pathway
Abstract
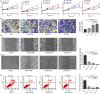
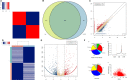
Methylation is the main form of RNA modification. N6-methyladenine (m6A) regulates the splicing and translation of mRNA. Alk B homologue 5 (ALKBH5) participates in the biological regulation of various cancers. However, its role in ovarian carcinogenesis has not been unveiled. In the present study, ALKBH5 showed higher expression in ovarian cancer tissue than in normal ovarian tissue, but lower expression in ovarian cancer cell lines than in normal ovarian cell lines. Interestingly, Toll-like receptor (TLR4), a molecular functioning in tumour microenvironment (TME), demonstrated the same expression trend. To investigate the effect of abnormal TME on ovarian carcinogenesis, we established an in vitro model in which macrophages and ovarian cancer cells were co-cultured. In the ovarian cancer cells co-cultured with M2 macrophages, the expression of ALKBH5 and TLR4 increased. We also verified that TLR4 up-regulated ALKBH5 expression via activating NF-κB pathway. Depending on transcriptome sequencing, m6A-Seq and m6A MeRIP, we found that NANOG served as a target in ALKBH5-mediated m6A modification. NANOG expression increased after mRNA demethylation, consequently enhancing the aggressiveness of ovarian cancer cells. In conclusion, highly expressed TLR4 activated NF-κB pathway, up-regulated ALKBH5 expression and increased m6A level and NANOG expression, all contributing to ovarian carcinogenesis. Our study revealed the role of m6A in ovarian carcinogenesis, providing a clue for inventing new target therapy.
Keywords: ALKBH5; RNA methylation; ovarian cancer; tumour microenvironment.
© 2020 The Authors. Journal of Cellular and Molecular Medicine published by Foundation for Cellular and Molecular Medicine and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare that they have no conflict interest in the study or preparation of the manuscript.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

