The Architecture of SARS-CoV-2 Transcriptome
- PMID: 32330414
- PMCID: PMC7179501
- DOI: 10.1016/j.cell.2020.04.011
The Architecture of SARS-CoV-2 Transcriptome
Abstract
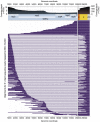
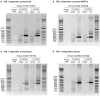
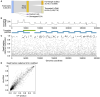
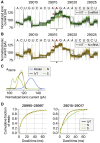
SARS-CoV-2 is a betacoronavirus responsible for the COVID-19 pandemic. Although the SARS-CoV-2 genome was reported recently, its transcriptomic architecture is unknown. Utilizing two complementary sequencing techniques, we present a high-resolution map of the SARS-CoV-2 transcriptome and epitranscriptome. DNA nanoball sequencing shows that the transcriptome is highly complex owing to numerous discontinuous transcription events. In addition to the canonical genomic and 9 subgenomic RNAs, SARS-CoV-2 produces transcripts encoding unknown ORFs with fusion, deletion, and/or frameshift. Using nanopore direct RNA sequencing, we further find at least 41 RNA modification sites on viral transcripts, with the most frequent motif, AAGAA. Modified RNAs have shorter poly(A) tails than unmodified RNAs, suggesting a link between the modification and the 3' tail. Functional investigation of the unknown transcripts and RNA modifications discovered in this study will open new directions to our understanding of the life cycle and pathogenicity of SARS-CoV-2.
Keywords: COVID-19; RNA modification; SARS-CoV-2; coronavirus; direct RNA sequencing; discontinuous transcription; epitranscriptome; nanopore; poly(A) tail; transcriptome.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests The authors declare no competing interests.
Figures







References
-
- Garalde D.R., Snell E.A., Jachimowicz D., Sipos B., Lloyd J.H., Bruce M., Pantic N., Admassu T., James P., Warland A. Highly parallel direct RNA sequencing on an array of nanopores. Nat. Methods. 2018;15:201–206. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

