Deficiency in the autophagy modulator Dram1 exacerbates pyroptotic cell death of Mycobacteria-infected macrophages
- PMID: 32332700
- PMCID: PMC7181687
- DOI: 10.1038/s41419-020-2477-1
Deficiency in the autophagy modulator Dram1 exacerbates pyroptotic cell death of Mycobacteria-infected macrophages
Abstract
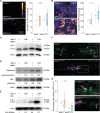
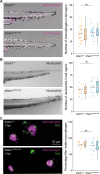
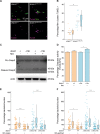
DNA damage regulated autophagy modulator 1 (DRAM1) is a stress-inducible regulator of autophagy and cell death. DRAM1 has been implicated in cancer, myocardial infarction, and infectious diseases, but the molecular and cellular functions of this transmembrane protein remain poorly understood. Previously, we have proposed DRAM1 as a host resistance factor for tuberculosis (TB) and a potential target for host-directed anti-infective therapies. In this study, we generated a zebrafish dram1 mutant and investigated its loss-of-function effects during Mycobacterium marinum (Mm) infection, a widely used model in TB research. In agreement with previous knockdown analysis, dram1 mutation increased the susceptibility of zebrafish larvae to Mm infection. RNA sequencing revealed major effects of Dram1 deficiency on metabolic, immune response, and cell death pathways during Mm infection, and only minor effects on proteinase and metabolic pathways were found under uninfected conditions. Furthermore, unchallenged dram1 mutants did not display overt autophagic defects, but autophagic targeting of Mm was reduced in the absence of Dram1. The phagocytic ability of macrophages in dram1 mutants was unaffected, but acidification of Mm-containing vesicles was strongly reduced, indicating that Dram1 is required for phagosome maturation. By in vivo imaging, we observed that Dram1-deficient macrophages fail to restrict Mm during early stages of infection. The resulting increase in bacterial burden could be reverted by knockdown of inflammatory caspase a (caspa) and gasdermin Eb (gsdmeb), demonstrating pyroptosis as the mechanism underlying premature cell death of Mm-infected macrophages in dram1 mutants. Collectively, these data demonstrate that dissemination of mycobacterial infection in zebrafish larvae is promoted in the absence of Dram1 due to reduced maturation of mycobacteria-containing vesicles, failed intracellular containment, and consequent pyroptotic death of infected macrophages. These results provide new evidence that Dram1 plays a central role in host resistance to intracellular infection, acting at the crossroad of autophagy and cell death.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

