Mapping the spreading routes of lymphatic metastases in human colorectal cancer
- PMID: 32332722
- PMCID: PMC7181746
- DOI: 10.1038/s41467-020-15886-6
Mapping the spreading routes of lymphatic metastases in human colorectal cancer
Abstract
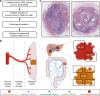
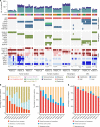
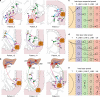
Lymphatic metastases are closely associated with tumor relapse and reduced survival in colorectal cancer (CRC). How tumor cells disseminate within the lymphatic network remains largely unknown. Here, we analyze the subclonal structure of 94 tumor samples, covering the primary tumors, lymph node metastases (LNMs), and liver metastases from 10 CRC patients. We portray a high-resolution lymphatic metastatic map for CRC by dividing LNMs into paracolic, intermediate, and central subgroups. Among the 61 metastatic routes identified, 38 (62.3%) are initiated from the primary tumors, 22 (36.1%) from LNMs, and 1 from liver metastasis (1.6%). In 5 patients, we find 6 LNMs that reseed 2 or more LNMs. We summarize 3 diverse modes of metastasis in CRC and show that skip spreading of tumor cells within the lymphatic network is common. Our study sheds light on the complicated metastatic pattern in CRC and has great clinical implications.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

