Chemical modifications of adenine base editor mRNA and guide RNA expand its application scope
- PMID: 32332735
- PMCID: PMC7181807
- DOI: 10.1038/s41467-020-15892-8
Chemical modifications of adenine base editor mRNA and guide RNA expand its application scope
Abstract
CRISPR-Cas9-associated base editing is a promising tool to correct pathogenic single nucleotide mutations in research or therapeutic settings. Efficient base editing requires cellular exposure to levels of base editors that can be difficult to attain in hard-to-transfect cells or in vivo. Here we engineer a chemically modified mRNA-encoded adenine base editor that mediates robust editing at various cellular genomic sites together with moderately modified guide RNA, and show its therapeutic potential in correcting pathogenic single nucleotide mutations in cell and animal models of diseases. The optimized chemical modifications of adenine base editor mRNA and guide RNA expand the applicability of CRISPR-associated gene editing tools in vitro and in vivo.
Conflict of interest statement
D.R.L. is a consultant and co-founder of Editas Medicine, Pairwise Plants, Prime Medicine, and Beam Therapeutics, companies that use genome editing. W.X. is a consultant for the Cystic Fibrosis Foundation Therapeutics Lab. The other authors declare no competing interests.
Figures
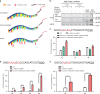
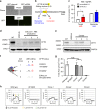
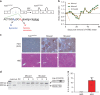
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

