RT-LAMP for rapid diagnosis of coronavirus SARS-CoV-2
- PMID: 32333644
- PMCID: PMC7264870
- DOI: 10.1111/1751-7915.13586
RT-LAMP for rapid diagnosis of coronavirus SARS-CoV-2
Abstract
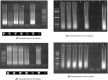
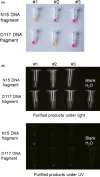
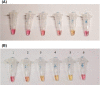
The pandemic coronavirus SARS-CoV-2 in the world has caused a large infected population suffering from COVID-19. To curb the spreading of the virus, WHO urgently demanded an extension of screening and testing; thus, a rapid and simple diagnostic method is needed. We applied a reverse transcription-loop-mediated isothermal amplification (RT-LAMP) to achieve the detection of SARS-CoV-2 in 30 min. We designed four sets of LAMP primers (6 primers in each set), targeting the viral RNA of SARS-CoV-2 in the regions of orf1ab, S gene and N gene. A colorimetric change was used to report the results, which enables the outcome of viral RNA amplification to be read by the naked eye without the need of expensive or dedicated instrument. The sensitivity can be 80 copies of viral RNA per ml in a sample. We validated the RT-LAMP method in a hospital in China, employing 16 clinic samples with 8 positives and 8 negatives. The testing results are consistent with the conventional RT-qPCR. In addition, we also show that one-step process without RNA extraction is feasible to achieve RNA amplification directly from a sample. This rapid, simple and sensitive RT-LAMP method paves a way for a large screening at public domain and hospitals, particularly regional hospitals and medical centres in rural areas.
© 2020 The Authors. Microbial Biotechnology published by John Wiley & Sons Ltd and Society for Applied Microbiology.
Conflict of interest statement
None declared.
Figures



References
-
- Boehme, C.C. , Nabeta, P. , Henostroza, G. , Raqib, R. , Rahim, Z. , Gerhardt, M. , et al (2007) Operational feasibility of using loop‐mediated isothermal amplification for diagnosis of pulmonary tuberculosis in microscopy centers of developing countries. J Clin Microbiol 45: 1936–1940. - PMC - PubMed
-
- Drosten, C. , Gottig, S. , Schilling, S. , Asper, M. , Panning, M. , Schmitz, H. , and Gunther, S. (2002) Rapid detection and quantification of RNA of Ebola and Marburg viruses, Lassa virus, Crimean‐Congo hemorrhagic fever virus, Rift Valley fever virus, Dengue virus, and Yellow fever virus by real‐time reverse transcription‐PCR. J Clin Microbiol 40: 2323–2330. - PMC - PubMed
-
- Hsieh, K. , Mage, P.L. , Csordas, A.T. , Eisenstein, M. , and Soh, H.T. (2014) Simultaneous elimination of carryover contamination and detection of DNA with uracil‐DNA‐glycosylase‐supplemented loop‐mediated isothermal amplification (UDG‐LAMP). Chem Commun 50: 3747–3749. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

