Deciphering MET-dependent modulation of global cellular responses to DNA damage by quantitative phosphoproteomics
- PMID: 32336009
- PMCID: PMC7266272
- DOI: 10.1002/1878-0261.12696
Deciphering MET-dependent modulation of global cellular responses to DNA damage by quantitative phosphoproteomics
Abstract
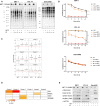
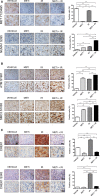
Increasing evidence suggests that interference with growth factor receptor tyrosine kinase (RTK) signaling can affect DNA damage response (DDR) networks, with a consequent impact on cellular responses to DNA-damaging agents widely used in cancer treatment. In that respect, the MET RTK is deregulated in abundance and/or activity in a variety of human tumors. Using two proteomic techniques, we explored how disrupting MET signaling modulates global cellular phosphorylation response to ionizing radiation (IR). Following an immunoaffinity-based phosphoproteomic discovery survey, we selected candidate phosphorylation sites for extensive characterization by targeted proteomics focusing on phosphorylation sites in both signaling networks. Several substrates of the DDR were confirmed to be modulated by sequential MET inhibition and IR, or MET inhibition alone. Upon combined treatment, for two substrates, NUMA1 S395 and CHEK1 S345, the gain and loss of phosphorylation, respectively, were recapitulated using invivo tumor models by immunohistochemistry, with possible utility in future translational research. Overall, we have corroborated phosphorylation sites at the intersection between MET and the DDR signaling networks, and suggest that these represent a class of proteins at the interface between oncogene-driven proliferation and genomic stability.
Keywords: ATM; DNA damage response; MET; ionizing radiation; mass spectrometry; receptor tyrosine kinase.
© 2020 The Authors. Published by FEBS Press and John Wiley & Sons Ltd.
Conflict of interest statement
A. Blaukat is listed as a co‐inventor on all patents related to Merck's c‐Met inhibitor listed in this manuscript. No potential conflicts of interest were disclosed by the other authors.
Figures


References
-
- Al‐Ejeh F, Kumar R, Wiegmans A, Lakhani SR, Brown MP and Khanna KK (2010) Harnessing the complexity of DNA‐damage response pathways to improve cancer treatment outcomes. Oncogene 29, 6085–6098. - PubMed
-
- Baschnagel AM, Williams L, Hanna A, Chen PY, Krauss DJ, Pruetz BL, Akervall J and Wilson GD (2014) c‐Met expression is a marker of poor prognosis in patients with locally advanced head and neck squamous cell carcinoma treated with chemoradiation. Int J Radiat Oncol Biol Phys 88, 701–707. - PubMed
-
- Benjamini Y and Hochberg Y (1995) Controlling the false discovery rate ‐ a practical and powerful approach to multiple testing. J Roy Stat Soc B Met 57, 289–300.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

