The dynamic interplay of host and viral enzymes in type III CRISPR-mediated cyclic nucleotide signalling
- PMID: 32338598
- PMCID: PMC7213978
- DOI: 10.7554/eLife.55852
The dynamic interplay of host and viral enzymes in type III CRISPR-mediated cyclic nucleotide signalling
Abstract
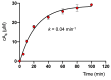
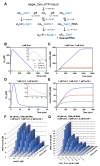
Cyclic nucleotide second messengers are increasingly implicated in prokaryotic anti-viral defence systems. Type III CRISPR systems synthesise cyclic oligoadenylate (cOA) upon detecting foreign RNA, activating ancillary nucleases that can be toxic to cells, necessitating mechanisms to remove cOA in systems that operate via immunity rather than abortive infection. Previously, we demonstrated that the Sulfolobus solfataricus type III-D CRISPR complex generates cyclic tetra-adenylate (cA4), activating the ribonuclease Csx1, and showed that subsequent RNA cleavage and dissociation acts as an 'off-switch' for the cyclase activity. Subsequently, we identified the cellular ring nuclease Crn1, which slowly degrades cA4 to reset the system (Rouillon et al., 2018), and demonstrated that viruses can subvert type III CRISPR immunity by means of a potent anti-CRISPR ring nuclease variant AcrIII-1. Here, we present a comprehensive analysis of the dynamic interplay between these enzymes, governing cyclic nucleotide levels and infection outcomes in virus-host conflict.
Keywords: CRISPR; E. coli; Sulfolobus solfataricus; biochemistry; chemical biology; cyclic oligoadenylate; ribonuclease; ring nuclease.
© 2020, Athukoralage et al.
Conflict of interest statement
JA, SG, CR, SG, CC, MW No competing interests declared
Figures











References
-
- Andrade WA, Firon A, Schmidt T, Hornung V, Fitzgerald KA, Kurt-Jones EA, Trieu-Cuot P, Golenbock DT, Kaminski PA. Group B Streptococcus degrades Cyclic-di-AMP to modulate STING-Dependent type I interferon production. Cell Host & Microbe. 2016;20:49–59. doi: 10.1016/j.chom.2016.06.003. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

