A tudor domain protein, SIMR-1, promotes siRNA production at piRNA-targeted mRNAs in C. elegans
- PMID: 32338603
- PMCID: PMC7255803
- DOI: 10.7554/eLife.56731
A tudor domain protein, SIMR-1, promotes siRNA production at piRNA-targeted mRNAs in C. elegans
Abstract
piRNAs play a critical role in the regulation of transposons and other germline genes. In Caenorhabditis elegans, regulation of piRNA target genes is mediated by the mutator complex, which synthesizes high levels of siRNAs through the activity of an RNA-dependent RNA polymerase. However, the steps between mRNA recognition by the piRNA pathway and siRNA amplification by the mutator complex are unknown. Here, we identify the Tudor domain protein, SIMR-1, as acting downstream of piRNA production and upstream of mutator complex-dependent siRNA biogenesis. Interestingly, SIMR-1 also localizes to distinct subcellular foci adjacent to P granules and Mutator foci, two phase-separated condensates that are the sites of piRNA-dependent mRNA recognition and mutator complex-dependent siRNA amplification, respectively. Thus, our data suggests a role for multiple perinuclear condensates in organizing the piRNA pathway and promoting mRNA regulation by the mutator complex.
Keywords: C. elegans; RNA silencing; chromosomes; gene expression; germ granules; germline; nuage; piRNAs; siRNAs.
Plain language summary
In the biological world, a process known as RNA interference helps cells to switch genes on and off and to defend themselves against harmful genetic material. This mechanism works by deactivating RNA sequences, the molecular templates cells can use to create proteins. Overall, RNA interference relies on the cell creating small RNA molecules that can target and inhibit the harmful RNA sequences that need to be silenced. More precisely, in round worms such as Caenorhabditis elegans, RNA interference happens in two steps. First, primary small RNAs identify the target sequences, which are then combatted by newly synthetised, secondary small RNAs. A number of proteins are also involved in both steps of the process. RNA interference is particularly important to preserve fertility, guarding sex cells against ‘rogue’ segments of genetic information that could be passed on to the next generation. In future sex cells, the proteins involved in RNA interference cluster together, forming a structure called a germ granule. Yet, little is known about the roles and identity of these proteins. To fill this knowledge gap, Manage et al. focused on the second stage of the RNA interference pathway in the germ granules of C. elegans, examining the molecules that physically interact with a key protein. This work revealed a new protein called SIMR-1. Looking into the role of SIMR-1 showed that the protein is required to amplify secondary small RNAs, but not to identify target sequences. However, it only promotes the creation of secondary small RNAs if a specific subtype of primary small RNAs have recognized the target RNAs for silencing. Further experiments also showed that within the germ granule, SIMR-1 is present in a separate substructure different from any compartment previously identified. This suggests that each substep of the RNA interference process takes place at a different location in the granule. In both C. elegans and humans, disruptions in the RNA interference pathway can lead to conditions such as cancer or infertility. Dissecting the roles of the proteins involved in this process in roundworms may help to better grasp how this process unfolds in mammals, and how it could be corrected in the case of disease.
© 2020, Manage et al.
Conflict of interest statement
KM, AR, DW, CU, DA, DN, KA, KB, RC, Bd, RK, TM, CP No competing interests declared
Figures

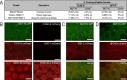

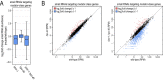
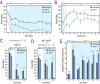
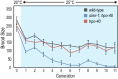



References
-
- Andralojc KM, Campbell AC, Kelly AL, Terrey M, Tanner PC, Gans IM, Senter-Zapata MJ, Khokhar ES, Updike DL. ELLI-1, a novel germline protein, modulates RNAi activity and P-granule accumulation in Caenorhabditis elegans. PLOS Genetics. 2017;13:e1006611. doi: 10.1371/journal.pgen.1006611. - DOI - PMC - PubMed
-
- Ashe A, Sapetschnig A, Weick EM, Mitchell J, Bagijn MP, Cording AC, Doebley AL, Goldstein LD, Lehrbach NJ, Le Pen J, Pintacuda G, Sakaguchi A, Sarkies P, Ahmed S, Miska EA. piRNAs can trigger a multigenerational epigenetic memory in the germline of C. elegans. Cell. 2012;150:88–99. doi: 10.1016/j.cell.2012.06.018. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

