Drug Research Meets Network Science: Where Are We?
- PMID: 32338900
- PMCID: PMC8007104
- DOI: 10.1021/acs.jmedchem.9b01989
Drug Research Meets Network Science: Where Are We?
Abstract
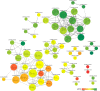
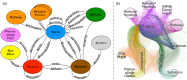
Network theory provides one of the most potent analysis tools for the study of complex systems. In this paper, we illustrate the network-based perspective in drug research and how it is coherent with the new paradigm of drug discovery. We first present data sources from which networks are built, then show some examples of how the networks can be used to investigate drug-related systems. A section is devoted to network-based inference applications, i.e., prediction methods based on interactomes, that can be used to identify putative drug-target interactions without resorting to 3D modeling. Finally, we present some aspects of Boolean networks dynamics, anticipating that it might become a very potent modeling framework to develop in silico screening protocols able to simulate phenotypic screening experiments. We conclude that network applications integrated with machine learning and 3D modeling methods will become an indispensable tool for computational drug discovery in the next years.
Conflict of interest statement
The authors declare no competing financial interest.
Figures


References
MeSH terms
LinkOut - more resources
Full Text Sources

