Targeted Killing of Pseudomonas aeruginosa by Pyocin G Occurs via the Hemin Transporter Hur
- PMID: 32339530
- PMCID: PMC7322526
- DOI: 10.1016/j.jmb.2020.04.020
Targeted Killing of Pseudomonas aeruginosa by Pyocin G Occurs via the Hemin Transporter Hur
Abstract
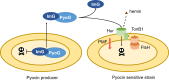
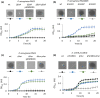
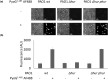
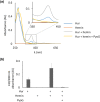
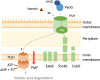
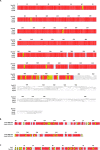
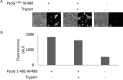
Pseudomonas aeruginosa is a priority pathogen for the development of new antibiotics, particularly because multi-drug-resistant strains of this bacterium cause serious nosocomial infections and are the leading cause of death in cystic fibrosis patients. Pyocins, bacteriocins of P. aeruginosa, are potent and diverse protein antibiotics that are deployed during bacterial competition. Pyocins are produced by more than 90% of P. aeruginosa strains and may have utility as last resort antibiotics against this bacterium. In this study, we explore the antimicrobial activity of a newly discovered pyocin called pyocin G (PyoG). We demonstrate that PyoG has broad killing activity against a collection of clinical P. aeruginosa isolates and is active in a Galleria mellonella infection model. We go on to identify cell envelope proteins that are necessary for the import of PyoG and its killing activity. PyoG recognizes bacterial cells by binding to Hur, an outer-membrane TonB-dependent transporter. Both pyocin and Hur interact with TonB1, which in complex with ExbB-ExbD links the proton motive force generated across the inner membrane with energy-dependent pyocin translocation across the outer membrane. Inner-membrane translocation of PyoG is dependent on the conserved inner-membrane AAA+ ATPase/protease, FtsH. We also report a functional exploration of the PyoG receptor. We demonstrate that Hur can bind to hemin in vitro and that this interaction is blocked by PyoG, confirming the role of Hur in hemin acquisition.
Keywords: FtsH; TonB-dependent transporter; bacteriocin; protein antibiotic; protein import.
Copyright © 2020 The Authors. Published by Elsevier Ltd.. All rights reserved.
Figures






References
-
- WHO . Media Centre; News Release: 2017. WHO Publishes List of Bacteria for which New Antibiotics Are Urgently Needed.http://www.who.int/mediacentre/news/releases/2017/bacteria-antibiotics-n... Available from:
-
- Kleanthous C. Swimming against the tide: progress and challenges in our understanding of colicin translocation. Nat. Rev. Microbiol. 2010;8:843–848. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

