DNA Damage/Repair Management in Cancers
- PMID: 32340362
- PMCID: PMC7226105
- DOI: 10.3390/cancers12041050
DNA Damage/Repair Management in Cancers
Abstract
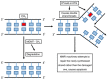
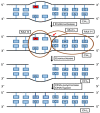
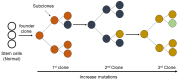
DNA damage is well recognized as a critical factor in cancer development and progression. DNA lesions create an abnormal nucleotide or nucleotide fragment, causing a break in one or both chains of the DNA strand. When DNA damage occurs, the possibility of generated mutations increases. Genomic instability is one of the most important factors that lead to cancer development. DNA repair pathways perform the essential role of correcting the DNA lesions that occur from DNA damaging agents or carcinogens, thus maintaining genomic stability. Inefficient DNA repair is a critical driving force behind cancer establishment, progression and evolution. A thorough understanding of DNA repair mechanisms in cancer will allow for better therapeutic intervention. In this review we will discuss the relationship between DNA damage/repair mechanisms and cancer, and how we can target these pathways.
Keywords: DNA damage; DNA lesion; DNA repair pathway; genomic instability.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

