Allelic polymorphism shapes community function in evolving Pseudomonas aeruginosa populations
- PMID: 32341475
- PMCID: PMC7368067
- DOI: 10.1038/s41396-020-0652-0
Allelic polymorphism shapes community function in evolving Pseudomonas aeruginosa populations
Abstract
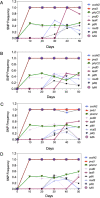
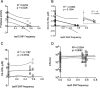
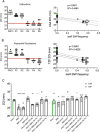
Pseudomonas aeruginosa is an opportunistic pathogen that chronically infects the lungs of individuals with cystic fibrosis (CF) by forming antibiotic-resistant biofilms. Emergence of phenotypically diverse isolates within CF P. aeruginosa populations has previously been reported; however, the impact of heterogeneity on social behaviors and community function is poorly understood. Here we describe how this heterogeneity impacts on behavioral traits by evolving the strain PAO1 in biofilms grown in a synthetic sputum medium for 50 days. We measured social trait production and antibiotic tolerance, and used a metagenomic approach to analyze and assess genomic changes over the duration of the evolution experiment. We found that (i) evolutionary trajectories were reproducible in independently evolving populations; (ii) over 60% of genomic diversity occurred within the first 10 days of selection. We then focused on quorum sensing (QS), a well-studied P. aeruginosa trait that is commonly mutated in strains isolated from CF lungs. We found that at the population level, (i) evolution in sputum medium selected for decreased the production of QS and QS-dependent traits; (ii) there was a significant correlation between lasR mutant frequency, the loss of protease, and the 3O-C12-HSL signal, and an increase in resistance to clinically relevant β-lactam antibiotics, despite no previous antibiotic exposure. Overall, our findings provide insights into the effect of allelic polymorphism on community functions in diverse P. aeruginosa populations. Further, we demonstrate that P. aeruginosa population and evolutionary dynamics can impact on traits important for virulence and can lead to increased tolerance to β-lactam antibiotics.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

