Genome-wide association meta-analyses combining multiple risk phenotypes provide insights into the genetic architecture of cutaneous melanoma susceptibility
- PMID: 32341527
- PMCID: PMC7255059
- DOI: 10.1038/s41588-020-0611-8
Genome-wide association meta-analyses combining multiple risk phenotypes provide insights into the genetic architecture of cutaneous melanoma susceptibility
Abstract
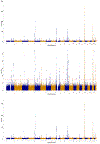
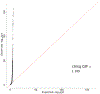
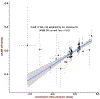
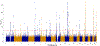
Most genetic susceptibility to cutaneous melanoma remains to be discovered. Meta-analysis genome-wide association study (GWAS) of 36,760 cases of melanoma (67% newly genotyped) and 375,188 controls identified 54 significant (P < 5 × 10-8) loci with 68 independent single nucleotide polymorphisms. Analysis of risk estimates across geographical regions and host factors suggests the acral melanoma subtype is uniquely unrelated to pigmentation. Combining this meta-analysis with GWAS of nevus count and hair color, and transcriptome association approaches, uncovered 31 potential secondary loci for a total of 85 cutaneous melanoma susceptibility loci. These findings provide insights into cutaneous melanoma genetic architecture, reinforcing the importance of nevogenesis, pigmentation and telomere maintenance, together with identifying potential new pathways for cutaneous melanoma pathogenesis.
Figures



References
-
- Secretan B et al. WHO International Agency for Research on Cancer Monograph Working Group A review of human carcinogens—Part E: tobacco, areca nut, alcohol, coal smoke, and salted fish. Lancet Oncol. 10, 1033–1034 (2009). - PubMed
-
- Ford D et al. Risk of cutaneous melanoma associated with a family history of the disease. Int. J. Cancer 62, 377–381 (1995). - PubMed
-
- Olsen CM, Carroll HJ & Whiteman DC Familial melanoma: a meta-analysis and estimates of attributable fraction. Cancer Epidemiol. Biomarkers Prev 19, 65–73 (2010). - PubMed
-
- Olsen CM, Carroll HJ & Whiteman DC Estimating the attributable fraction for melanoma: a meta-analysis of pigmentary characteristics and freckling. Int. J. Cancer 127, 2430–2445 (2010). - PubMed
Methods References
-
- DerSimonian R & Laird N Meta-analysis in clinical trials. Control. Clin. Trials 7, 177–188 (1986). - PubMed
Publication types
MeSH terms
Grants and funding
- MC_UU_00007/10/MRC_/Medical Research Council/United Kingdom
- ZIA CP010200/ImNIH/Intramural NIH HHS/United States
- P01 CA087969/CA/NCI NIH HHS/United States
- 29186/CRUK_/Cancer Research UK/United Kingdom
- C490/A16561/CRUK_/Cancer Research UK/United Kingdom
- R01 CA122838/CA/NCI NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- R01 CA049449/CA/NCI NIH HHS/United States
- U01 CA049449/CA/NCI NIH HHS/United States
- R01 CA100264/CA/NCI NIH HHS/United States
- R01 CA133996/CA/NCI NIH HHS/United States
- R01 CA083115/CA/NCI NIH HHS/United States
- P01 CA055075/CA/NCI NIH HHS/United States
- WT084766/Z/08/Z/WT_/Wellcome Trust/United Kingdom
- P30 CA008748/CA/NCI NIH HHS/United States
- 19167/CRUK_/Cancer Research UK/United Kingdom
- R01 CA088363/CA/NCI NIH HHS/United States
- U19 CA148112/CA/NCI NIH HHS/United States
- 10118/CRUK_/Cancer Research UK/United Kingdom
- 10589/CRUK_/Cancer Research UK/United Kingdom
LinkOut - more resources
Full Text Sources
Medical

