Systematic comparison of single-cell and single-nucleus RNA-sequencing methods
- PMID: 32341560
- PMCID: PMC7289686
- DOI: 10.1038/s41587-020-0465-8
Systematic comparison of single-cell and single-nucleus RNA-sequencing methods
Erratum in
-
Author Correction: Systematic comparison of single-cell and single-nucleus RNA-sequencing methods.Nat Biotechnol. 2020 Jun;38(6):756. doi: 10.1038/s41587-020-0534-z. Nat Biotechnol. 2020. PMID: 32341567
Abstract
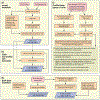
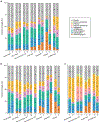
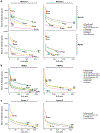
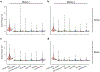
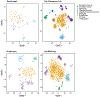
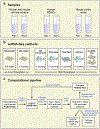
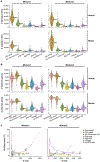
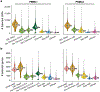
The scale and capabilities of single-cell RNA-sequencing methods have expanded rapidly in recent years, enabling major discoveries and large-scale cell mapping efforts. However, these methods have not been systematically and comprehensively benchmarked. Here, we directly compare seven methods for single-cell and/or single-nucleus profiling-selecting representative methods based on their usage and our expertise and resources to prepare libraries-including two low-throughput and five high-throughput methods. We tested the methods on three types of samples: cell lines, peripheral blood mononuclear cells and brain tissue, generating 36 libraries in six separate experiments in a single center. To directly compare the methods and avoid processing differences introduced by the existing pipelines, we developed scumi, a flexible computational pipeline that can be used with any single-cell RNA-sequencing method. We evaluated the methods for both basic performance, such as the structure and alignment of reads, sensitivity and extent of multiplets, and for their ability to recover known biological information in the samples.
Conflict of interest statement
COMPETING FINANCIAL INTERESTS
A.R. is a founder and equity holder in Celsius Therapeutics, an equity holder in Immunitas, and an SAB member of Syros Pharmaceuticals, Neogene Therapeutics, and Thermo Fisher Scientific. A.K.S. is a founder of, and consultant for, Honeycomb Biotechnologies, Inc. which manufactures Seq-Well peripherals. A.K.S. and A.R. are also named inventors on patents filed by the Broad Institute related to either Drop-seq (AR and AKS), DroNc-seq (A.R.), or Seq-Well (A.K.S). The interests of A.K.S. and A.R. were reviewed and are subject to a management plan overseen by their institutions in accordance with their conflict of interest policies. The other authors declare no competing financial interests.
Figures






References
METHODS-ONLY REFERENCES
-
- Liao Y, Smyth GK & Shi W featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

