Zebrafish B cell acute lymphoblastic leukemia: new findings in an old model
- PMID: 32341750
- PMCID: PMC7170496
- DOI: 10.18632/oncotarget.27555
Zebrafish B cell acute lymphoblastic leukemia: new findings in an old model
Abstract
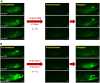
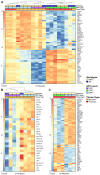
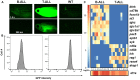
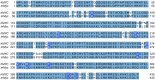
Acute lymphoblastic leukemia (ALL) is the most common pediatric, and ninth most common adult, cancer. ALL can develop in either B or T lymphocytes, but B-lineage ALL (B-ALL) exceeds T-ALL clinically. As for other cancers, animal models allow study of the molecular mechanisms driving ALL. Several zebrafish (Danio rerio) T-ALL models have been reported, but until recently, robust D. rerio B-ALL models were not described. Then, D. rerio B-ALL was discovered in two related zebrafish transgenic lines; both were already known to develop T-ALL. Here, we report new B-ALL findings in one of these models, fish expressing transgenic human MYC (hMYC). We describe B-ALL incidence in a large cohort of hMYC fish, and show B-ALL in two new lines where T-ALL does not interfere with B-ALL detection. We also demonstrate B-ALL responses to steroid and radiation treatments, which effect ALL remissions, but are usually followed by prompt relapses. Finally, we report gene expression in zebrafish B lymphocytes and B-ALL, in both bulk samples and single B- and T-ALL cells. Using these gene expression profiles, we compare differences between the two new D. rerio B-ALL models, which are both driven by transgenic mammalian MYC oncoproteins. Collectively, these new data expand the utility of this new vertebrate B-ALL model.
Keywords: ALL; MYC; acute lymphoblastic leukemia; lymphocyte; zebrafish.
Conflict of interest statement
CONFLICTS OF INTEREST Authors declare no conflicts of interests.
Figures


Similar articles
-
Isolating Malignant and Non-Malignant B Cells from lck:eGFP Zebrafish.J Vis Exp. 2019 Feb 22;(144). doi: 10.3791/59191. J Vis Exp. 2019. PMID: 30855581
-
Heritable T-cell malignancy models established in a zebrafish phenotypic screen.Leukemia. 2009 Oct;23(10):1825-35. doi: 10.1038/leu.2009.116. Epub 2009 Jun 11. Leukemia. 2009. PMID: 19516274 Free PMC article.
-
Isolation of the Side Population in Myc-induced T-cell Acute Lymphoblastic Leukemia in Zebrafish.J Vis Exp. 2017 May 4;(123):55711. doi: 10.3791/55711. J Vis Exp. 2017. PMID: 28518092 Free PMC article.
-
Zebrafish models of leukemia.Methods Cell Biol. 2017;138:563-592. doi: 10.1016/bs.mcb.2016.11.013. Epub 2017 Jan 7. Methods Cell Biol. 2017. PMID: 28129858 Review.
-
Tackling Acute Lymphoblastic Leukemia-One Fish at a Time.Int J Mol Sci. 2019 Oct 25;20(21):5313. doi: 10.3390/ijms20215313. Int J Mol Sci. 2019. PMID: 31731471 Free PMC article. Review.
Cited by
-
Zebrafish disease models in drug discovery: from preclinical modelling to clinical trials.Nat Rev Drug Discov. 2021 Aug;20(8):611-628. doi: 10.1038/s41573-021-00210-8. Epub 2021 Jun 11. Nat Rev Drug Discov. 2021. PMID: 34117457 Free PMC article. Review.
-
Genetic Engineering of Zebrafish in Cancer Research.Cancers (Basel). 2020 Aug 4;12(8):2168. doi: 10.3390/cancers12082168. Cancers (Basel). 2020. PMID: 32759814 Free PMC article. Review.
-
Diverse Epithelial Lymphocytes in Zebrafish Revealed Using a Novel Scale Biopsy Method.J Immunol. 2024 Dec 15;213(12):1902-1914. doi: 10.4049/jimmunol.2300818. J Immunol. 2024. PMID: 39503619 Free PMC article.
-
Dynamic Changes in Lymphocyte Populations Establish Zebrafish as a Thymic Involution Model.J Immunol. 2024 Jun 1;212(11):1733-1743. doi: 10.4049/jimmunol.2300495. J Immunol. 2024. PMID: 38656392 Free PMC article.
-
IRF4 drives clonal evolution and lineage choice in a zebrafish model of T-cell lymphoma.Nat Commun. 2022 May 3;13(1):2420. doi: 10.1038/s41467-022-30053-9. Nat Commun. 2022. PMID: 35504924 Free PMC article.
References
-
- Pui CH, Yang JJ, Hunger SP, Pieters R, Schrappe M, Biondi A, Vora A, Baruchel A, Silverman LB, Schmiegelow K, Escherich G, Horibe K, Benoit YC, et al.. Childhood Acute Lymphoblastic Leukemia: Progress Through Collaboration. J Clin Oncol. 2015; 33:2938–48. 10.1200/JCO.2014.59.1636. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

