Light-Activatable, 2,5-Disubstituted Tetrazoles for the Proteome-wide Profiling of Aspartates and Glutamates in Living Bacteria
- PMID: 32342004
- PMCID: PMC7181327
- DOI: 10.1021/acscentsci.9b01268
Light-Activatable, 2,5-Disubstituted Tetrazoles for the Proteome-wide Profiling of Aspartates and Glutamates in Living Bacteria
Abstract
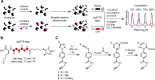
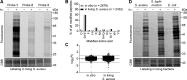
Covalent inhibitors have recently seen a resurgence of interest in drug development. Nevertheless, compounds, which do not rely on an enzymatic activity, have almost exclusively been developed to target cysteines. Expanding the scope to other amino acids would be largely facilitated by the ability to globally monitor their engagement by covalent inhibitors. Here, we present the use of light-activatable 2,5-disubstituted tetrazoles that allow quantifying 8971 aspartates and glutamates in the bacterial proteome with excellent selectivity. Using these probes, we competitively map the binding sites of two isoxazolium salts and introduce hydrazonyl chlorides as a new class of carboxylic-acid-directed covalent protein ligands. As the probes are unreactive prior to activation, they allow global profiling even in living Gram-positive and Gram-negative bacteria. Taken together, this method to monitor aspartates and glutamates proteome-wide will lay the foundation to efficiently develop covalent inhibitors targeting these amino acids.
Copyright © 2020 American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures


References
-
- Hendlin D.; Stapley E. O.; Jackson M.; Wallick H.; Miller A. K.; Wolf F. J.; Miller T. W.; Chaiet L.; Kahan F. M.; Foltz E. L.; Woodruff H. B.; Mata J. M.; Hernandez S.; Mochales S. Phosphonomycin, a new antibiotic produced by strains of streptomyces. Science 1969, 166, 122–123. 10.1126/science.166.3901.122. - DOI - PubMed
-
- Nishimura H.; Mayama M.; Komatsu Y.; Kato H.; Shimaoka N.; Tanaka Y. Showdomycin, a New Antibiotic from a Streptomyces Sp. J. Antibiot. 1964, 17, 148–155. - PubMed
-
- Smith P. A.; Koehler M. F. T.; Girgis H. S.; Yan D.; Chen Y.; Chen Y.; Crawford J. J.; Durk M. R.; Higuchi R. I.; Kang J.; Murray J.; Paraselli P.; Park S.; Phung W.; Quinn J. G.; Roberts T. C.; Rouge L.; Schwarz J. B.; Skippington E.; Wai J.; Xu M.; Yu Z.; Zhang H.; Tan M. W.; Heise C. E. Optimized arylomycins are a new class of Gram-negative antibiotics. Nature 2018, 561, 189–194. 10.1038/s41586-018-0483-6. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

