RT-qPCR Testing of SARS-CoV-2: A Primer
- PMID: 32344568
- PMCID: PMC7215906
- DOI: 10.3390/ijms21083004
RT-qPCR Testing of SARS-CoV-2: A Primer
Abstract
Testing for the presence of coronavirus is an essential diagnostic tool for monitoring and managing the current COVID-19 pandemic. The only reliable test in current use for testing acute infection targets the genome of SARS-CoV-2, and the most widely used method is quantitative fluorescence-based reverse transcription polymerase chain reaction (RT-qPCR). Despite its ubiquity, there is a significant amount of uncertainty about how this test works, potential throughput and reliability. This has resulted in widespread misrepresentation of the problems faced using this test during the current COVID-19 epidemic. This primer provides simple, straightforward and impartial information about RT-qPCR.
Keywords: COVID-19; SARS; pandemic; real-time fluorescence PCR; reverse transcription.
Conflict of interest statement
The authors declare no conflict of interest.
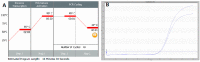
Figures


References
-
- Bustin S.A. A-Z of Quantitative PCR. IUL Press; La Jolla, CA, USA: 2004.
-
- Bustin S., Benes V., Garson J.A., Hellemans J., Huggett J., Kubista M., Mueller R., Nolan T., Pfaffl M., Shipley G.L., et al. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin. Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

