The F-box protein FBXL16 up-regulates the stability of C-MYC oncoprotein by antagonizing the activity of the F-box protein FBW7
- PMID: 32345600
- PMCID: PMC7278360
- DOI: 10.1074/jbc.RA120.012658
The F-box protein FBXL16 up-regulates the stability of C-MYC oncoprotein by antagonizing the activity of the F-box protein FBW7
Abstract
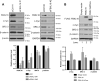
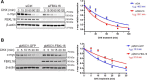
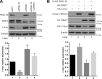
F-box proteins, such as F-box/WD repeat-containing protein 7 (FBW7), are essential components of the SKP1-CUL1-F-box (SCF) E3 ubiquitin ligases. They bind to S-phase kinase-associated protein 1 (SKP1) through the F-box motif and deliver their protein substrate to the E3 ligase complex for ubiquitination and subsequent degradation. F-box and leucine-rich repeat protein 16 (FBXL16) is a poorly studied F-box protein. Because it does not interact with the scaffold protein cullin 1 (CUL1), we hypothesized that FBXL16 might not form a functional SCF-E3 ligase complex. In the present study, we found that FBXL16 up-regulates the levels of proteins targeted by SCF-E3 ligases, such as C-MYC, β-catenin, and steroid receptor coactivator 3 (SRC-3). Focusing on C-MYC, a well-known oncoprotein overexpressed in most human cancers, we show that FBXL16 stabilizes C-MYC by antagonizing FBW7-mediated C-MYC ubiquitination and degradation. Further, we found that, although FBXL16 does not interact with CUL1, it interacts with SKP1 via its N-terminal F-box domain and with its substrate C-MYC via its C-terminal leucine-rich repeats (LRRs) domain. We found that both the F-box domain and the LRR domain are important for FBXL16-mediated C-MYC stabilization. In line with its role in up-regulating the levels of the C-MYC and SRC-3 oncoproteins, FBXL16 promoted cancer cell growth and migration and colony formation in soft agar. Our findings reveal that FBXL16 is an F-box protein that antagonizes the activity of another F-box protein, FBW7, and thereby increases C-MYC stability, resulting in increased cancer cell growth and invasiveness.
Keywords: E3 ubiquitin ligase; F-box and leucine-rich repeat protein 16 (FBXL16); F-box protein; F-box/WD repeat-containing protein 7 (FBW7); Myc (c-Myc); cancer; cell migration; protein homeostasis; protein stability; proto-oncogene C-Myc (C-MYC); ubiquitylation (ubiquitination).
© 2020 Morel et al.
Conflict of interest statement
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
Figures


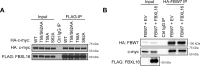
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

