A Family of Argonaute-Interacting Proteins Gates Nuclear RNAi
- PMID: 32348780
- PMCID: PMC7613089
- DOI: 10.1016/j.molcel.2020.04.007
A Family of Argonaute-Interacting Proteins Gates Nuclear RNAi
Abstract
Nuclear RNA interference (RNAi) pathways work together with histone modifications to regulate gene expression and enact an adaptive response to transposable RNA elements. In the germline, nuclear RNAi can lead to trans-generational epigenetic inheritance (TEI) of gene silencing. We identified and characterized a family of nuclear Argonaute-interacting proteins (ENRIs) that control the strength and target specificity of nuclear RNAi in C. elegans, ensuring faithful inheritance of epigenetic memories. ENRI-1/2 prevent misloading of the nuclear Argonaute NRDE-3 with small RNAs that normally effect maternal piRNAs, which prevents precocious nuclear translocation of NRDE-3 in the early embryo. Additionally, they are negative regulators of nuclear RNAi triggered from exogenous sources. Loss of ENRI-3, an unstable protein expressed mostly in the male germline, misdirects the RNAi response to transposable elements and impairs TEI. The ENRIs determine the potency and specificity of nuclear RNAi responses by gating small RNAs into specific nuclear Argonautes.
Keywords: 22G-RNA; Argonaute; NRDE-3; RNA interference; RNAi; piRNA; trans-generational epigenetic inheritance.
Copyright © 2020. Published by Elsevier Inc.
Conflict of interest statement
Declaration of Interests E.A.M. is a founder and director of STORM Therapeutics. This company has not contributed to this research in any way. The authors declare no competing interests.
Figures
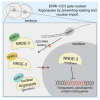



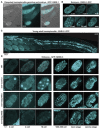
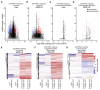
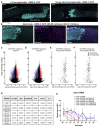

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

