Macromolecular crowding links ribosomal protein gene dosage to growth rate in Vibrio cholerae
- PMID: 32349767
- PMCID: PMC7191768
- DOI: 10.1186/s12915-020-00777-5
Macromolecular crowding links ribosomal protein gene dosage to growth rate in Vibrio cholerae
Abstract
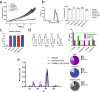
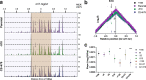
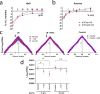
Background: In fast-growing bacteria, the genomic location of ribosomal protein (RP) genes is biased towards the replication origin (oriC). This trait allows optimizing their expression during exponential phase since oriC neighboring regions are in higher dose due to multifork replication. Relocation of s10-spc-α locus (S10), which codes for most of the RP, to ectopic genomic positions shows that its relative distance to the oriC correlates to a reduction on its dosage, its expression, and bacterial growth rate. However, a mechanism linking S10 dosage to cell physiology has still not been determined.
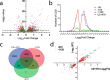
Results: We hypothesized that S10 dosage perturbations impact protein synthesis capacity. Strikingly, we observed that in Vibrio cholerae, protein production capacity was independent of S10 position. Deep sequencing revealed that S10 relocation altered chromosomal replication dynamics and genome-wide transcription. Such changes increased as a function of oriC-S10 distance. Since RP constitutes a large proportion of cell mass, lower S10 dosage could lead to changes in macromolecular crowding, impacting cell physiology. Accordingly, cytoplasm fluidity was higher in mutants where S10 is most distant from oriC. In hyperosmotic conditions, when crowding differences are minimized, the growth rate and replication dynamics were highly alleviated in these strains.
Conclusions: The genomic location of RP genes ensures its optimal dosage. However, besides of its essential function in translation, their genomic position sustains an optimal macromolecular crowding essential for maximizing growth. Hence, this could be another mechanism coordinating DNA replication to bacterial growth.
Keywords: Bacterial chromosome; Bacterial physiology; Growth rate; Macromolecular crowding; Ribosomal proteins; Synthetic biology; Vibrio cholerae.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
- ANR-10-BLAN-131301/Agence Nationale de la Recherche/International
- ANR-14-CE10-0007/Agence Nationale de la Recherche/International
- ANR-10-LABX-62-IBEID/Agence Nationale de la Recherche/International
- PICT-2017-0424 to ASB/Agencia Nacional de Promoción Científica y Tecnológica/International
- EMBO-ALTF-1473-2010/European Molecular Biology Organization/International
LinkOut - more resources
Full Text Sources

