Synthetic hybrids of six yeast species
- PMID: 32350251
- PMCID: PMC7190663
- DOI: 10.1038/s41467-020-15559-4
Synthetic hybrids of six yeast species
Abstract
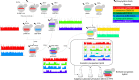
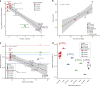
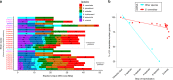
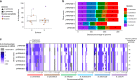
Allopolyploidy generates diversity by increasing the number of copies and sources of chromosomes. Many of the best-known evolutionary radiations, crops, and industrial organisms are ancient or recent allopolyploids. Allopolyploidy promotes differentiation and facilitates adaptation to new environments, but the tools to test its limits are lacking. Here we develop an iterative method of Hybrid Production (iHyPr) to combine the genomes of multiple budding yeast species, generating Saccharomyces allopolyploids of at least six species. When making synthetic hybrids, chromosomal instability and cell size increase dramatically as additional copies of the genome are added. The six-species hybrids initially grow slowly, but they rapidly regain fitness and adapt, even as they retain traits from multiple species. These new synthetic yeast hybrids and the iHyPr method have potential applications for the study of polyploidy, genome stability, chromosome segregation, and bioenergy.
Conflict of interest statement
The Wisconsin Alumni Research Foundation has filed a patent application entitled “Synthetic yeast cells and methods of making and using same” (describing the HyPr and iHyPr methods with W.G.A., D.P., and C.T.H. as inventors, US application #15/805950, filed 11/7/2017, pending). K.J.F., R.V.M., M.G.B., E.J.U., and R.L.W. declare no competing interests.
Figures


References
-
- Mable BK, Alexandrou MA, Taylor MI. Genome duplication in amphibians and fish: an extended synthesis. J. Zool. 2011;284:151–182. doi: 10.1111/j.1469-7998.2011.00829.x. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

