Advanced magnetic resonance spectroscopic neuroimaging: Experts' consensus recommendations
- PMID: 32350978
- PMCID: PMC7606742
- DOI: 10.1002/nbm.4309
Advanced magnetic resonance spectroscopic neuroimaging: Experts' consensus recommendations
Abstract
Magnetic resonance spectroscopic imaging (MRSI) offers considerable promise for monitoring metabolic alterations associated with disease or injury; however, to date, these methods have not had a significant impact on clinical care, and their use remains largely confined to the research community and a limited number of clinical sites. The MRSI methods currently implemented on clinical MRI instruments have remained essentially unchanged for two decades, with only incremental improvements in sequence implementation. During this time, a number of technological developments have taken place that have already greatly benefited the quality of MRSI measurements within the research community and which promise to bring advanced MRSI studies to the point where the technique becomes a true imaging modality, while making the traditional review of individual spectra a secondary requirement. Furthermore, the increasing use of biomedical MR spectroscopy studies has indicated clinical areas where advanced MRSI methods can provide valuable information for clinical care. In light of this rapidly changing technological environment and growing understanding of the value of MRSI studies for biomedical studies, this article presents a consensus from a group of experts in the field that reviews the state-of-the-art for clinical proton MRSI studies of the human brain, recommends minimal standards for further development of vendor-provided MRSI implementations, and identifies areas which need further technical development.
Keywords: brain; magnetic resonance spectroscopic imaging; review.
© 2020 John Wiley & Sons, Ltd.
Figures
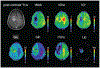


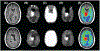



References
-
- Barker PB, Bizzi A, De Stefano N, Gullapalli R, Lin DDM. Clinical MR spectroscopy: Techniques and applications. Cambridge, UK: Cambridge University Press; 2009.
-
- Maudsley AA, Hilal SK, Perman WH, Simon HE. Spatially resolved high-resolution spectroscopy by "four-dimensional" NMR. J Magn Reson. 1983;51: 147–152.
-
- Mansfield P Spatial-mapping of the chemical-shift in NMR. J Phys D Appl Phys. 1983;16:L235–L238. - PubMed
Publication types
MeSH terms
Grants and funding
- R01 MH118695/MH/NIMH NIH HHS/United States
- R01 EB016064/EB/NIBIB NIH HHS/United States
- P30 GM122734/GM/NIGMS NIH HHS/United States
- R21 CA241714/CA/NCI NIH HHS/United States
- R01 EB006841/EB/NIBIB NIH HHS/United States
- R01 CA172210/CA/NCI NIH HHS/United States
- P 30701/FWF_/Austrian Science Fund FWF/Austria
- P50 CA097257/CA/NCI NIH HHS/United States
- R01 CA127612/CA/NCI NIH HHS/United States
- P50 CA165962/CA/NCI NIH HHS/United States
- R01 EB020407/EB/NIBIB NIH HHS/United States
- R01 CA200808/CA/NCI NIH HHS/United States
- P01 CA118816/CA/NCI NIH HHS/United States
- R01 CA211080/CA/NCI NIH HHS/United States
- KLI 718/FWF_/Austrian Science Fund FWF/Austria
LinkOut - more resources
Full Text Sources

