Genomic Investigation of the Strawberry Pathogen Phytophthora fragariae Indicates Pathogenicity Is Associated With Transcriptional Variation in Three Key Races
- PMID: 32351458
- PMCID: PMC7174552
- DOI: 10.3389/fmicb.2020.00490
Genomic Investigation of the Strawberry Pathogen Phytophthora fragariae Indicates Pathogenicity Is Associated With Transcriptional Variation in Three Key Races
Abstract
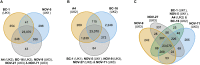
The oomycete Phytophthora fragariae is a highly destructive pathogen of cultivated strawberry (Fragaria × ananassa), causing the root rotting disease, "red core". The host-pathogen interaction has a well described gene-for-gene resistance relationship, but to date neither candidate avirulence nor resistance genes have been identified. We sequenced a set of American, Canadian, and United Kingdom isolates of known race type, along with three representatives of the closely related pathogen of the raspberry (Rubus idaeus), P. rubi, and found a clear population structure, with a high degree of nucleotide divergence seen between some race types and abundant private variation associated with race types 4 and 5. In contrast, between isolates defined as United Kingdom races 1, 2, and 3 (UK1-2-3) there was no evidence of gene loss or gain; or the presence of insertions/deletions (INDELs) or Single Nucleotide Polymorphisms (SNPs) within or in proximity to putative pathogenicity genes could be found associated with race variation. Transcriptomic analysis of representative UK1-2-3 isolates revealed abundant expression variation in key effector family genes associated with pathogen race; however, further long read sequencing did not reveal any long range polymorphisms to be associated with avirulence to race UK2 or UK3 resistance, suggesting either control in trans or other stable forms of epigenetic modification modulating gene expression. This work reveals the combined power of population resequencing to uncover race structure in pathosystems and in planta transcriptomic analysis to identify candidate avirulence genes. This work has implications for the identification of putative avirulence genes in the absence of associated expression data and points toward the need for detailed molecular characterisation of mechanisms of effector regulation and silencing in oomycete plant pathogens.
Keywords: RNA-Seq; host–microbe interactions; nanopore sequencing; oomycete; population resequencing; race structure; red core.
Copyright © 2020 Adams, Armitage, Sobczyk, Bates, Tabima, Kronmiller, Tyler, Grünwald, Dunwell, Nellist and Harrison.
Figures




References
-
- Adams T. M. (2019). Pathogenomics of Phytophthora fragariae, the Causal Agent of Strawberry Red Core Disease. Ph.D. thesis, University of Reading, Reading.
-
- Aronesty E. (2013). Comparison of sequencing utility programs. Open Bioinform. J. 7 1–8. 10.2174/1875036201307010001 - DOI
LinkOut - more resources
Full Text Sources

