The Biotherapeutic Potential of Lactobacillus reuteri Characterized Using a Target-Specific Selection Process
- PMID: 32351460
- PMCID: PMC7176361
- DOI: 10.3389/fmicb.2020.00532
The Biotherapeutic Potential of Lactobacillus reuteri Characterized Using a Target-Specific Selection Process
Abstract
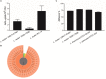
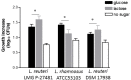
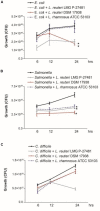
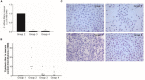
A growing body of clinical and experimental data supports the view that the efficacy of probiotics is strain-specific and restricted to particular pathological conditions, which means that newly isolated probiotic strains need to be targeted to a specific disease. Following national and international guidelines, we used a conventional in vitro experimental approach to characterize a novel strain of Lactobacillus reuteri, LMG P-27481, for safety (sensitivity to antibiotics and genome analysis) and putative efficacy (resistance to gastro-intestinal transit, adhesiveness, induction of cytokines, and release of antimicrobial metabolites). In vitro assays, which were carried out to examine the probiotic's effect on diarrhea (lactose utilization, inhibition of pathogens such as bacteria and Rotavirus), showed that it was more efficacious with respect to well-known reference strains in antagonizing Clostridioides difficile (CD). Data confirming that the probiotic can effectively treat CD colitis was gained from in vivo trials involving mice conditioned with large spectrum antibiotics before they were subjected to CD challenge. Two out of the three antibiotic-treated groups received daily LMG P-27481 for different time durations in order to simulate a preventive approach (LMG P-27481 administered prior to CD challenge) or an antagonistic one (LMG P-27481 administered after CD challenge). Both approaches significantly reduced, with respect to the untreated controls, CD DNA concentrations in caecum and C. difficile toxin titers in the gut lumen. In addition, LMG P-27481 supplementation significantly mitigated body weight loss and the extent of inflammatory infiltrate and tissue damage. The study results, which need to be confirmed by in vivo clinical trials, have demonstrated that the L. reuteri LMG P-27481 strain is a promising probiotic candidate for the treatment of CD infection.
Keywords: diarrhea; gut; intestine; lactobacilli; pathogen; probiotic.
Copyright © 2020 Sagheddu, Uggeri, Belogi, Remollino, Brun, Bernabè, Moretti, Porzionato, Morelli, Castagliuolo and Elli.
Figures



References
-
- Ausiello C. M., Cerquetti M., Fedele G., Spensieri F., Palazzo R., Nasso M., et al. (2006). Surface layer proteins from Clostridium difficile induce inflammatory and regulatory cytokines in human monocytes and dendritic cells. Microbes Infect. 8 2640–2646. 10.1016/j.micinf.2006.07.009 - DOI - PubMed
LinkOut - more resources
Full Text Sources

