Quantitative Study of the Chiral Organization of the Phage Genome Induced by the Packaging Motor
- PMID: 32353255
- PMCID: PMC7203069
- DOI: 10.1016/j.bpj.2020.03.030
Quantitative Study of the Chiral Organization of the Phage Genome Induced by the Packaging Motor
Abstract
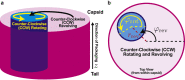
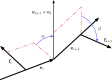
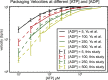
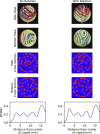
Molecular motors that translocate DNA are ubiquitous in nature. During morphogenesis of double-stranded DNA bacteriophages, a molecular motor drives the viral genome inside a protein capsid. Several models have been proposed for the three-dimensional geometry of the packaged genome, but very little is known of the signature of the molecular packaging motor. For instance, biophysical experiments show that in some systems, DNA rotates during the packaging reaction, but most current biophysical models fail to incorporate this property. Furthermore, studies including rotation mechanisms have reached contradictory conclusions. In this study, we compare the geometrical signatures imposed by different possible mechanisms for the packaging motors: rotation, revolution, and rotation with revolution. We used a previously proposed kinetic Monte Carlo model of the motor, combined with Brownian dynamics simulations of DNA to simulate deterministic and stochastic motor models. We find that rotation is necessary for the accumulation of DNA writhe and for the chiral organization of the genome. We observe that although in the initial steps of the packaging reaction, the torsional strain of the genome is released by rotation of the molecule, in the later stages, it is released by the accumulation of writhe. We suggest that the molecular motor plays a key role in determining the final structure of the encapsidated genome in bacteriophages.
Copyright © 2020 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures


References
-
- Casjens S.R. The DNA-packaging nanomotor of tailed bacteriophages. Nat. Rev. Microbiol. 2011;9:647–657. - PubMed
-
- Kellenberger E., Carlemalm E., De Haller G. Considerations on the condensation and the degree of compactness in non-eukaryotic DNA-containing plasmas. In: Gualerzi C.O., Pon C.L., editors. Bacterial Chromatin. Springer; 1986. pp. 11–25.
-
- Leforestier A., Brasilès S., Livolant F. Bacteriophage T5 DNA ejection under pressure. J. Mol. Biol. 2008;384:730–739. - PubMed
-
- Panja D., Molineux I.J. Dynamics of bacteriophage genome ejection in vitro and in vivo. Phys. Biol. 2010;7:045006. - PubMed

