Co-evolution of mutagenic genome editors and vertebrate adaptive immunity
- PMID: 32353821
- PMCID: PMC7768089
- DOI: 10.1016/j.coi.2020.03.001
Co-evolution of mutagenic genome editors and vertebrate adaptive immunity
Abstract
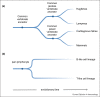
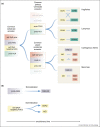
The adaptive immune systems of all vertebrates rely on self-DNA mutating enzymes to assemble their antigen receptors in lymphocytes of their two principal lineages. In jawed vertebrates, the RAG1/2 recombinase directs V(D)J recombination of B cell and T cell receptor genes, whereas the activation-induced cytidine deaminase AID engages in their secondary modification. The recombination activating genes (RAG) 1 and 2 evolved from an ancient transposon-encoded genome modifier into a self-DNA mutator serving adaptive immunity; this was possible as a result of domestication, involving several changes in RAG1 and RAG2 proteins suppressing transposition and instead facilitating-coupled cleavage and recombination. By contrast, recent evidence supports the notion that the antigen receptors of T-like and B-like cells of jawless vertebrates, designated variable lymphocyte receptors (VLRs), are somatically assembled through a process akin to gene conversion that is believed to be dependent on the activities of distant relatives of AID, the cytidine deaminases CDA1 and CDA2, respectively. It appears, therefore, that the precursors of AID and CDAs underwent a domestication process that changed their target range from foreign nucleic acids to self-DNA; this multi-step evolutionary process ensured that the threat to host genome integrity was minimized. Here, we review recent findings illuminating the evolutionary steps associated with the domestication of the two groups of genome editors, RAG1/2 and cytidine deaminases, indicating how they became the driving forces underlying the emergence of vertebrate adaptive immune systems.
Copyright © 2020 The Authors. Published by Elsevier Ltd.. All rights reserved.
Figures
References
-
- Liaw S.H., Chang Y.J., Lai C.T., Chang H.C., Chang G.G. Crystal structure of Bacillus subtilis guanine deaminase: the first domain-swapped structure in the cytidine deaminase superfamily. J Biol Chem. 2004;279:35479–35485. - PubMed
-
- Hamilton C.E., Papavasiliou F.N., Rosenberg B.R. Diverse functions for DNA and RNA editing in the immune system. RNA Biol. 2010;7:220–228. - PubMed
-
- Thomas C.M., Nielsen K.M. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat Rev Microbiol. 2005;3:711–721. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

