A SARS-CoV-2 protein interaction map reveals targets for drug repurposing
- PMID: 32353859
- PMCID: PMC7431030
- DOI: 10.1038/s41586-020-2286-9
A SARS-CoV-2 protein interaction map reveals targets for drug repurposing
Abstract
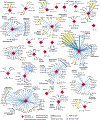
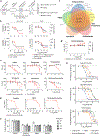
A newly described coronavirus named severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which is the causative agent of coronavirus disease 2019 (COVID-19), has infected over 2.3 million people, led to the death of more than 160,000 individuals and caused worldwide social and economic disruption1,2. There are no antiviral drugs with proven clinical efficacy for the treatment of COVID-19, nor are there any vaccines that prevent infection with SARS-CoV-2, and efforts to develop drugs and vaccines are hampered by the limited knowledge of the molecular details of how SARS-CoV-2 infects cells. Here we cloned, tagged and expressed 26 of the 29 SARS-CoV-2 proteins in human cells and identified the human proteins that physically associated with each of the SARS-CoV-2 proteins using affinity-purification mass spectrometry, identifying 332 high-confidence protein-protein interactions between SARS-CoV-2 and human proteins. Among these, we identify 66 druggable human proteins or host factors targeted by 69 compounds (of which, 29 drugs are approved by the US Food and Drug Administration, 12 are in clinical trials and 28 are preclinical compounds). We screened a subset of these in multiple viral assays and found two sets of pharmacological agents that displayed antiviral activity: inhibitors of mRNA translation and predicted regulators of the sigma-1 and sigma-2 receptors. Further studies of these host-factor-targeting agents, including their combination with drugs that directly target viral enzymes, could lead to a therapeutic regimen to treat COVID-19.
Figures












Update of
-
A SARS-CoV-2-Human Protein-Protein Interaction Map Reveals Drug Targets and Potential Drug-Repurposing.bioRxiv [Preprint]. 2020 Mar 27:2020.03.22.002386. doi: 10.1101/2020.03.22.002386. bioRxiv. 2020. Update in: Nature. 2020 Jul;583(7816):459-468. doi: 10.1038/s41586-020-2286-9. PMID: 32511329 Free PMC article. Updated. Preprint.
Comment in
-
A map of SARS-CoV-2 and host cell interactions.Nat Rev Immunol. 2020 Jun;20(6):351. doi: 10.1038/s41577-020-0318-1. Nat Rev Immunol. 2020. PMID: 32303698 Free PMC article. No abstract available.
-
Anti-viral properties of antipsychotic medications in the time of COVID-19.Psychiatry Res. 2021 Jan;295:113626. doi: 10.1016/j.psychres.2020.113626. Epub 2020 Nov 30. Psychiatry Res. 2021. PMID: 33290940 Free PMC article. No abstract available.
-
Olanzapine, risperidone and quetiapine: Do these atypical antipsychotics have a protective effect for SARS-CoV-2?Schizophr Res. 2022 Mar;241:140-141. doi: 10.1016/j.schres.2022.01.035. Epub 2022 Jan 24. Schizophr Res. 2022. PMID: 35123336 Free PMC article. No abstract available.
References
-
- Novel Coronavirus (2019-nCoV) situation reports. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situatio....
Publication types
MeSH terms
Substances
Grants and funding
- F32 GM133144/GM/NIGMS NIH HHS/United States
- R01 GM024485/GM/NIGMS NIH HHS/United States
- 1R01CA244550/NH/NIH HHS/United States
- T32 AI060537/AI/NIAID NIH HHS/United States
- R01 GM117189/GM/NIGMS NIH HHS/United States
- R01 AI122747/AI/NIAID NIH HHS/United States
- F32 CA239333/CA/NCI NIH HHS/United States
- R01 CA221969/CA/NCI NIH HHS/United States
- K08 HL124068/HL/NHLBI NIH HHS/United States
- 1F32CA236347-01/NH/NIH HHS/United States
- R35 GM122481/GM/NIGMS NIH HHS/United States
- T32 CA108462/CA/NCI NIH HHS/United States
- P01 AI063302/AI/NIAID NIH HHS/United States
- F30 CA239476/CA/NCI NIH HHS/United States
- P50 AI150476/AI/NIAID NIH HHS/United States
- R01 AI143292/AI/NIAID NIH HHS/United States
- F32 CA236347/CA/NCI NIH HHS/United States
- R01 GM140440/GM/NIGMS NIH HHS/United States
- HHSN272201400008C/AI/NIAID NIH HHS/United States
- R01 AI120694/AI/NIAID NIH HHS/United States
- T32 GM008284/GM/NIGMS NIH HHS/United States
- T32 GM067547/GM/NIGMS NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- U19 AI135990/AI/NIAID NIH HHS/United States
- R01 CA244550/CA/NCI NIH HHS/United States
- R01 AI118974/AI/NIAID NIH HHS/United States
- P30 CA023100/CA/NCI NIH HHS/United States
- R35 GM118099/GM/NIGMS NIH HHS/United States
- R01 HG009979/HG/NHGRI NIH HHS/United States
- P01 AI120943/AI/NIAID NIH HHS/United States
- P30 AI027763/AI/NIAID NIH HHS/United States
- U19 AI135972/AI/NIAID NIH HHS/United States
- T32 GM064337/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

