Community curation in PomBase: enabling fission yeast experts to provide detailed, standardized, sharable annotation from research publications
- PMID: 32353878
- PMCID: PMC7192550
- DOI: 10.1093/database/baaa028
Community curation in PomBase: enabling fission yeast experts to provide detailed, standardized, sharable annotation from research publications
Abstract
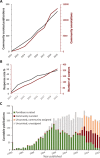
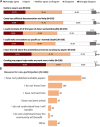
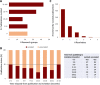
Maximizing the impact and value of scientific research requires efficient knowledge distribution, which increasingly depends on the integration of standardized published data into online databases. To make data integration more comprehensive and efficient for fission yeast research, PomBase has pioneered a community curation effort that engages publication authors directly in FAIR-sharing of data representing detailed biological knowledge from hypothesis-driven experiments. Canto, an intuitive online curation tool that enables biologists to describe their detailed functional data using shared ontologies, forms the core of PomBase's system. With 8 years' experience, and as the author response rate reaches 50%, we review community curation progress and the insights we have gained from the project. We highlight incentives and nudges we deploy to maximize participation, and summarize project outcomes, which include increased knowledge integration and dissemination as well as the unanticipated added value arising from co-curation by publication authors and professional curators.
© The Author(s) 2020. Published by Oxford University Press.
Figures

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

