Antimicrobial peptides: Application informed by evolution
- PMID: 32355003
- PMCID: PMC8097767
- DOI: 10.1126/science.aau5480
Antimicrobial peptides: Application informed by evolution
Abstract
Antimicrobial peptides (AMPs) are essential components of immune defenses of multicellular organisms and are currently in development as anti-infective drugs. AMPs have been classically assumed to have broad-spectrum activity and simple kinetics, but recent evidence suggests an unexpected degree of specificity and a high capacity for synergies. Deeper evaluation of the molecular evolution and population genetics of AMP genes reveals more evidence for adaptive maintenance of polymorphism in AMP genes than has previously been appreciated, as well as adaptive loss of AMP activity. AMPs exhibit pharmacodynamic properties that reduce the evolution of resistance in target microbes, and AMPs may synergize with one another and with conventional antibiotics. Both of these properties make AMPs attractive for translational applications. However, if AMPs are to be used clinically, it is crucial to understand their natural biology in order to lessen the risk of collateral harm and avoid the crisis of resistance now facing conventional antibiotics.
Copyright © 2020 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Conflict of interest statement
Figures

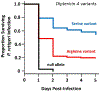
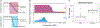

References
-
- Koo HB, Seo J, Antimicrobial peptides under clinical investigation. Pept. Sci 111, e24122 (2019). doi: 10.1002/pep2.24122 - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

