Development and validation of an interpretable deep learning framework for Alzheimer's disease classification
- PMID: 32357201
- PMCID: PMC7296847
- DOI: 10.1093/brain/awaa137
Development and validation of an interpretable deep learning framework for Alzheimer's disease classification
Abstract
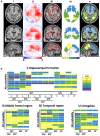
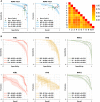
Alzheimer's disease is the primary cause of dementia worldwide, with an increasing morbidity burden that may outstrip diagnosis and management capacity as the population ages. Current methods integrate patient history, neuropsychological testing and MRI to identify likely cases, yet effective practices remain variably applied and lacking in sensitivity and specificity. Here we report an interpretable deep learning strategy that delineates unique Alzheimer's disease signatures from multimodal inputs of MRI, age, gender, and Mini-Mental State Examination score. Our framework linked a fully convolutional network, which constructs high resolution maps of disease probability from local brain structure to a multilayer perceptron and generates precise, intuitive visualization of individual Alzheimer's disease risk en route to accurate diagnosis. The model was trained using clinically diagnosed Alzheimer's disease and cognitively normal subjects from the Alzheimer's Disease Neuroimaging Initiative (ADNI) dataset (n = 417) and validated on three independent cohorts: the Australian Imaging, Biomarker and Lifestyle Flagship Study of Ageing (AIBL) (n = 382), the Framingham Heart Study (n = 102), and the National Alzheimer's Coordinating Center (NACC) (n = 582). Performance of the model that used the multimodal inputs was consistent across datasets, with mean area under curve values of 0.996, 0.974, 0.876 and 0.954 for the ADNI study, AIBL, Framingham Heart Study and NACC datasets, respectively. Moreover, our approach exceeded the diagnostic performance of a multi-institutional team of practicing neurologists (n = 11), and high-risk cerebral regions predicted by the model closely tracked post-mortem histopathological findings. This framework provides a clinically adaptable strategy for using routinely available imaging techniques such as MRI to generate nuanced neuroimaging signatures for Alzheimer's disease diagnosis, as well as a generalizable approach for linking deep learning to pathophysiological processes in human disease.
Keywords: Alzheimer’s disease; biomarkers; dementia; neurodegeneration; structural MRI.
© The Author(s) (2020). Published by Oxford University Press on behalf of the Guarantors of Brain.
Figures




References
-
- Barkhof F, Polvikoski TM, van Straaten EC, Kalaria RN, Sulkava R, Aronen HJ, et al.The significance of medial temporal lobe atrophy: a postmortem MRI study in the very old. Neurology 2007; 69: 1521–7. - PubMed
-
- Beekly DL, Ramos EM, van Belle G, Deitrich W, Clark AD, Jacka ME, et al.The National Alzheimer’s Coordinating Center (NACC) Database: an Alzheimer disease database. Alzheimer Dis Assoc Disord 2004; 18: 270–7. - PubMed
-
- Bohnen NI, Djang DS, Herholz K, Anzai Y, Minoshima S.. Effectiveness and safety of 18F-FDG PET in the evaluation of dementia: a review of the recent literature. J Nucl Med 2012; 53: 59–71. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AG054076/AG/NIA NIH HHS/United States
- R01 AG016495/AG/NIA NIH HHS/United States
- HHSN268201500001C/HL/NHLBI NIH HHS/United States
- RF1 AG062109/AG/NIA NIH HHS/United States
- P30 AG013846/AG/NIA NIH HHS/United States
- RF1 AG072654/AG/NIA NIH HHS/United States
- R01 HL159620/HL/NHLBI NIH HHS/United States
- HHSN268201500001I/HL/NHLBI NIH HHS/United States
- R56 AG062109/AG/NIA NIH HHS/United States
- U19 AG068753/AG/NIA NIH HHS/United States
- UL1 TR001430/TR/NCATS NIH HHS/United States
- P30 AG066546/AG/NIA NIH HHS/United States
- R01 AG033040/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Research Materials

