Comparative Transcriptomic Analysis to Identify the Genes Related to Delayed Gland Morphogenesis in Gossypium bickii
- PMID: 32357512
- PMCID: PMC7290383
- DOI: 10.3390/genes11050472
Comparative Transcriptomic Analysis to Identify the Genes Related to Delayed Gland Morphogenesis in Gossypium bickii
Abstract
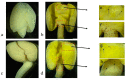
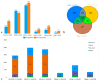
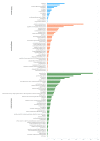
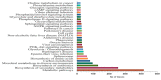
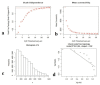
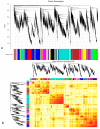
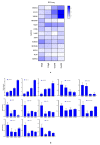
Cotton is one of the major industrial crops that supply natural fibers and oil for industries. This study was conducted to understand the mechanism of delayed gland morphogenesis in seeds of Gossypium bickii. In this study, we compared glandless seeds of G. bickii with glanded seeds of Gossypium arboreum. High-throughput sequencing technology was used to explore and classify the expression patterns of gland-related genes in seeds and seedlings of cotton plants. Approximately 131.33 Gigabases of raw data from 12 RNA sequencing samples with three biological replicates were generated. A total of 7196 differentially-expressed genes (DEGs) were identified in all transcriptome data. Among them, 3396 genes were found up-regulated and 3480 genes were down-regulated. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) annotations were performed to identify different functions between genes unique to glandless imbibed seeds and glanded seedlings. Co-expression network analysis revealed four modules that were identified as highly associated with the development of glandless seeds. Here the hub genes in each module were identified by weighted gene co-expression network analysis (WGCNA). In total, we have selected 13 genes involved in transcription factors, protein and MYB-related functions, that were differentially expressed in transcriptomic data and validated by quantitative reverse-transcription polymerase chain reaction (qRT-PCR). These selected genes may play an important role for delayed gland morphogenesis. Our study provides comprehensive insight into the key genes related to glandless traits of seeds and plants, and can be further exploited by functional and molecular studies.
Keywords: DEGs; Gossypium bickii; RNA-seq; WGCNA.
Conflict of interest statement
All the authors declare no conflict of interest.
Figures

References
-
- Fryxell P.A. A redefinition of the tribe Gossypieae. Bot. Gaz. 1968;129:296–308. doi: 10.1086/336448. - DOI
-
- Stanford E.E., Viehoever A. Chemistry and histology of the glands of the cotton plant, with notes on the occurrence of similar glands in related plants. J. Agric. Res. 1918;13:419–435.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

